
🐟 Weighted Average Detection Efficiency (WADE) Positioning Analysis
Positioning Animals with Acoustic Telemetry Using Detection Range Information - Simulation Application
Jake Brownscombe
2025-09-16
Source:vignettes/WADE_Simulation.Rmd
WADE_Simulation.RmdAnalysis Overview
This tutorial demonstrates the Weighted Average Detection Efficiency (WADE) approach for animal positioning using passive acoustic telemetry. It uses detection range information to generate estimates of probable animal locations based on detection patterns, which is useful for estimating space use (home range) and habitat selection. This approach using simulated data is a useful exercise to assess positioning accuracy and optimal settings relative to known tracks, which may be useful to conduct prior to field data applications where positioning can be influenced by system design, detection range, and animal behaviour.
📡 Receiver Array Design
Generate Optimal Array Configuration
# Generate regularly spaced receiver array
# Note: actual number may vary slightly from n_points to optimize spacing
points_regular <- generate_exact_regular_points(
depth_raster,
n_points = 80,
seed = 123
)
# Visualize array design
plot_points_on_input(depth_raster, points_regular) +
ggtitle("Receiver Array Design",
subtitle = paste("Regular spacing with", nrow(points_regular), "receivers")) +
theme_minimal()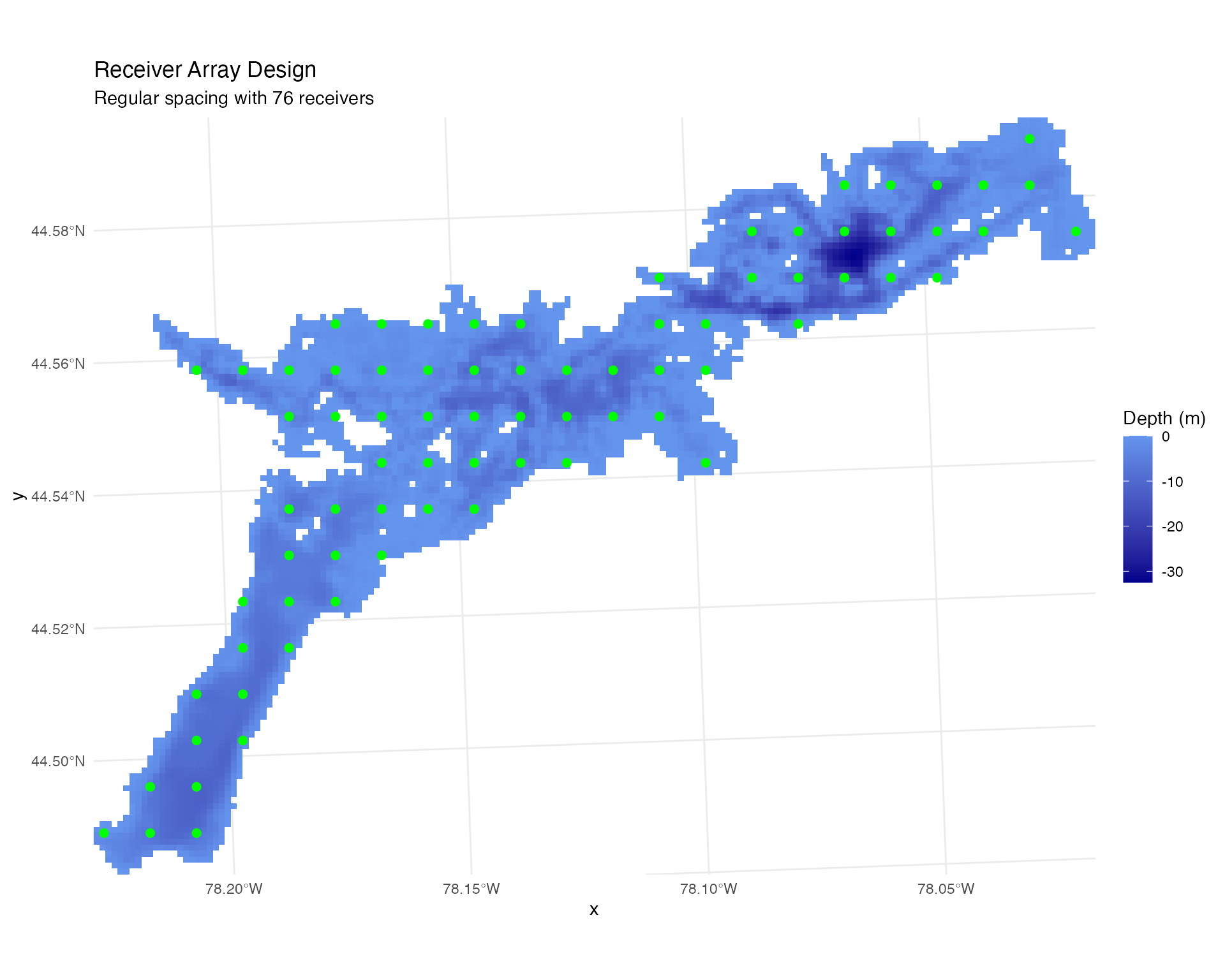
Calculate Station Distances
# Calculate cost-weighted distances between all grid cells and receivers
station_distances <- calculate_station_distances(
raster = depth_raster,
receiver_frame = points_regular,
max_distance = 30000, # Maximum distance to consider (30 km)
station_col = "station_id"
)## Creating transition matrix...
## Calculating distances for station 1 of 76 (ID: 1 )
## Calculating distances for station 2 of 76 (ID: 2 )
## Calculating distances for station 3 of 76 (ID: 3 )
## Calculating distances for station 4 of 76 (ID: 4 )
## Calculating distances for station 5 of 76 (ID: 5 )
## Calculating distances for station 6 of 76 (ID: 6 )
## Calculating distances for station 7 of 76 (ID: 7 )
## Calculating distances for station 8 of 76 (ID: 8 )
## Calculating distances for station 9 of 76 (ID: 9 )
## Calculating distances for station 10 of 76 (ID: 10 )
## Calculating distances for station 11 of 76 (ID: 11 )
## Calculating distances for station 12 of 76 (ID: 12 )
## Calculating distances for station 13 of 76 (ID: 13 )
## Calculating distances for station 14 of 76 (ID: 14 )
## Calculating distances for station 15 of 76 (ID: 15 )
## Calculating distances for station 16 of 76 (ID: 16 )
## Calculating distances for station 17 of 76 (ID: 17 )
## Calculating distances for station 18 of 76 (ID: 18 )
## Calculating distances for station 19 of 76 (ID: 19 )
## Calculating distances for station 20 of 76 (ID: 20 )
## Calculating distances for station 21 of 76 (ID: 21 )
## Calculating distances for station 22 of 76 (ID: 22 )
## Calculating distances for station 23 of 76 (ID: 23 )
## Calculating distances for station 24 of 76 (ID: 24 )
## Calculating distances for station 25 of 76 (ID: 25 )
## Calculating distances for station 26 of 76 (ID: 26 )
## Calculating distances for station 27 of 76 (ID: 27 )
## Calculating distances for station 28 of 76 (ID: 28 )
## Calculating distances for station 29 of 76 (ID: 29 )
## Calculating distances for station 30 of 76 (ID: 30 )
## Calculating distances for station 31 of 76 (ID: 31 )
## Calculating distances for station 32 of 76 (ID: 32 )
## Calculating distances for station 33 of 76 (ID: 33 )
## Calculating distances for station 34 of 76 (ID: 34 )
## Calculating distances for station 35 of 76 (ID: 35 )
## Calculating distances for station 36 of 76 (ID: 36 )
## Calculating distances for station 37 of 76 (ID: 37 )
## Calculating distances for station 38 of 76 (ID: 38 )
## Calculating distances for station 39 of 76 (ID: 39 )
## Calculating distances for station 40 of 76 (ID: 40 )
## Calculating distances for station 41 of 76 (ID: 41 )
## Calculating distances for station 42 of 76 (ID: 42 )
## Calculating distances for station 43 of 76 (ID: 43 )
## Calculating distances for station 44 of 76 (ID: 44 )
## Calculating distances for station 45 of 76 (ID: 45 )
## Calculating distances for station 46 of 76 (ID: 46 )
## Calculating distances for station 47 of 76 (ID: 47 )
## Calculating distances for station 48 of 76 (ID: 48 )
## Calculating distances for station 49 of 76 (ID: 49 )
## Calculating distances for station 50 of 76 (ID: 50 )
## Calculating distances for station 51 of 76 (ID: 51 )
## Calculating distances for station 52 of 76 (ID: 52 )
## Calculating distances for station 53 of 76 (ID: 53 )
## Calculating distances for station 54 of 76 (ID: 54 )
## Calculating distances for station 55 of 76 (ID: 55 )
## Calculating distances for station 56 of 76 (ID: 56 )
## Calculating distances for station 57 of 76 (ID: 57 )
## Calculating distances for station 58 of 76 (ID: 58 )
## Calculating distances for station 59 of 76 (ID: 59 )
## Calculating distances for station 60 of 76 (ID: 60 )
## Calculating distances for station 61 of 76 (ID: 61 )
## Calculating distances for station 62 of 76 (ID: 62 )
## Calculating distances for station 63 of 76 (ID: 63 )
## Calculating distances for station 64 of 76 (ID: 64 )
## Calculating distances for station 65 of 76 (ID: 65 )
## Calculating distances for station 66 of 76 (ID: 66 )
## Calculating distances for station 67 of 76 (ID: 67 )
## Calculating distances for station 68 of 76 (ID: 68 )
## Calculating distances for station 69 of 76 (ID: 69 )
## Calculating distances for station 70 of 76 (ID: 70 )
## Calculating distances for station 71 of 76 (ID: 71 )
## Calculating distances for station 72 of 76 (ID: 72 )
## Calculating distances for station 73 of 76 (ID: 73 )
## Calculating distances for station 74 of 76 (ID: 74 )
## Calculating distances for station 75 of 76 (ID: 75 )
## Calculating distances for station 76 of 76 (ID: 76 )
## Converting to long format...
## Distance calculations complete!
## Result contains 366168 station-cell combinations
## Station IDs ( integer ): 1, 2, 3, 4, 5 ...
str(station_distances) #contains shortest path, euclidean distances, and tortuosity ## tibble [366,168 × 8] (S3: tbl_df/tbl/data.frame)
## $ cell_id : int [1:366168] 156 157 158 159 160 321 322 323 324 325 ...
## $ x : num [1:366168] 735854 735954 736054 736154 736254 ...
## $ y : num [1:366168] 4941891 4941891 4941891 4941891 4941891 ...
## $ raster_value : num [1:366168] -0.03541 -0.25052 -0.85191 -0.4325 -0.00829 ...
## $ cost_distance : num [1:366168] 20488 20588 20688 20788 20888 ...
## $ station_no : int [1:366168] 1 1 1 1 1 1 1 1 1 1 ...
## $ straight_distance: num [1:366168] 19477 19556 19635 19715 19794 ...
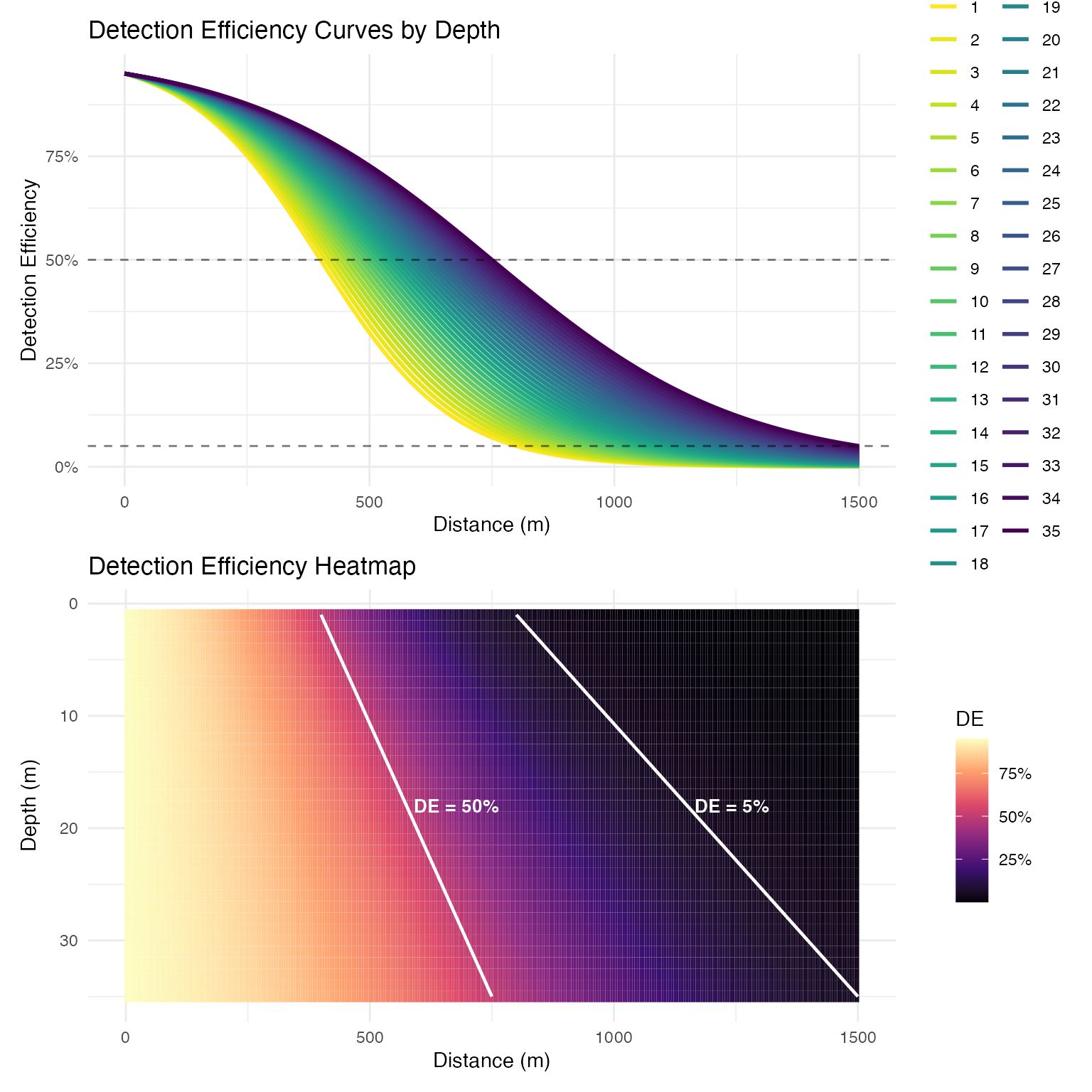
## $ tortuosity : num [1:366168] 1.05 1.05 1.05 1.05 1.06 ...📊 Detection Range Modeling
Create Depth-Dependent Detection Model
# Generate logistic detection efficiency model with depth-dependency
logistic_DE <- create_logistic_curve_depth(
min_depth = 1,
max_depth = 35,
d50_min_depth = 400, # 50% detection at 400m in shallow water
d95_min_depth = 800, # 95% detection at 800m in shallow water
d50_max_depth = 750, # 50% detection at 750m in deep water
d95_max_depth = 1500, # 95% detection at 1500m in deep water
plot = TRUE,
return_model = TRUE,
return_object = TRUE
)
##
## Call:
## stats::glm(formula = DE ~ dist_m * depth_m, family = stats::binomial(link = "logit"),
## data = predictions)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -0.124692 -0.019879 0.006263 0.023533 0.050386
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 2.825e+00 1.331e-01 21.228 <2e-16 ***
## dist_m -6.738e-03 2.256e-04 -29.871 <2e-16 ***
## depth_m 5.807e-03 6.209e-03 0.935 0.35
## dist_m:depth_m 8.277e-05 9.398e-06 8.807 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 6031.202 on 10534 degrees of freedom
## Residual deviance: 11.686 on 10531 degrees of freedom
## AIC: 4332.3
##
## Number of Fisher Scoring iterations: 6Apply Detection Model to System
# Predict detection efficiency for all receiver-cell combinations
station_distances$DE_pred <- stats::predict(
logistic_DE$log_model,
newdata = station_distances %>%
dplyr::rename(dist_m = cost_distance) %>%
dplyr::mutate(depth_m = abs(raster_value)),
type = "response"
)
# Visualize detection field for example station
ggplot(station_distances %>% dplyr::filter(station_no == 10),
aes(x, y, fill = DE_pred)) +
geom_raster() +
scale_fill_viridis_c(option = "magma", name = "Detection\nEfficiency") +
ggtitle("Detection Efficiency Field (Station 10)",
subtitle = "Probability of detection decreases with distance and depth") +
theme_minimal() +
coord_sf()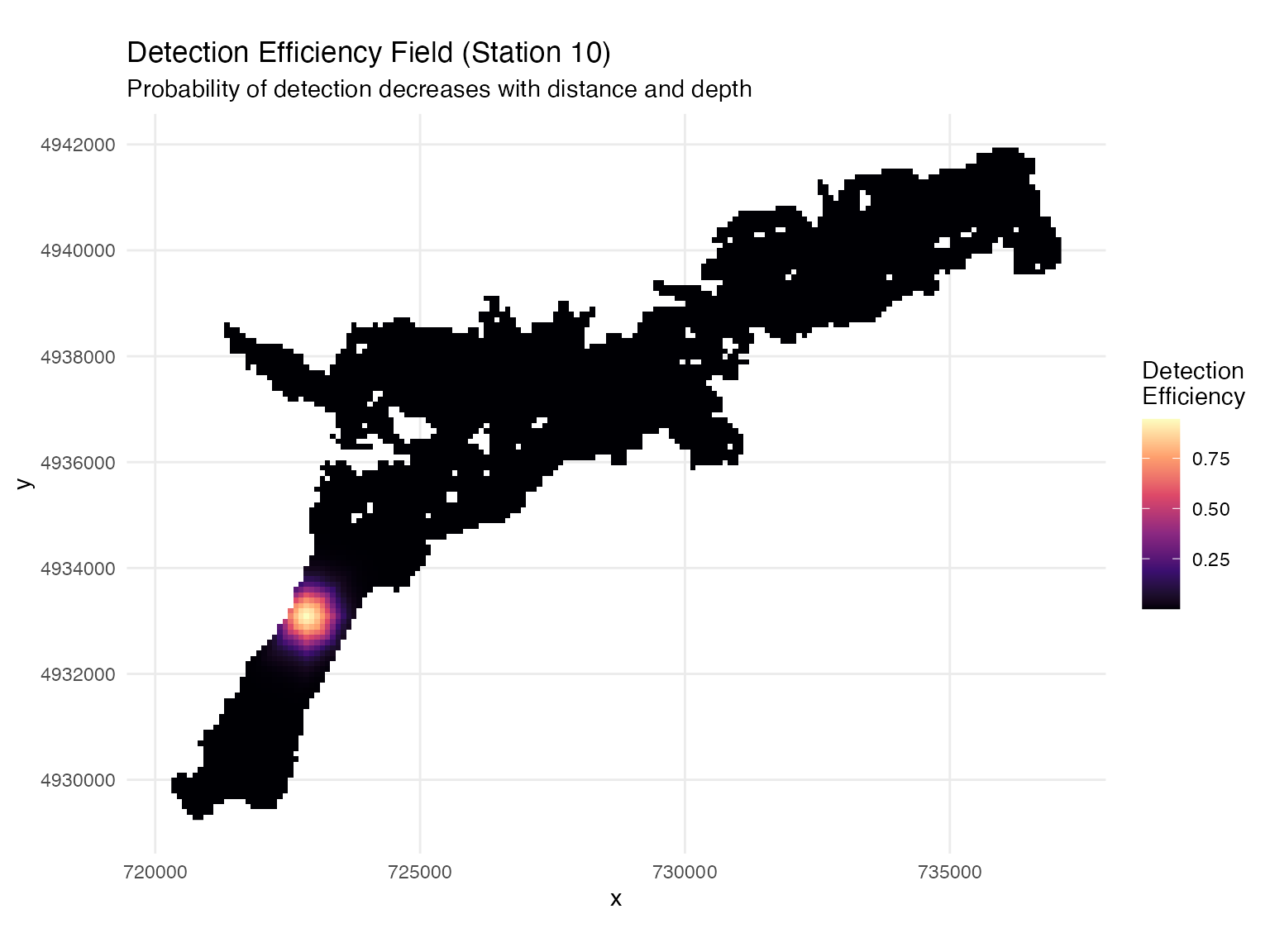
System-Wide Detection Coverage
# Calculate cumulative system detection efficiency
# This will be used for generating absence points later
system_DE <- calculate_detection_system(
distance_frame = station_distances,
receiver_frame = points_regular,
model = logistic_DE$log_model,
output_type = "cumulative", # Cumulative probability across all receivers
plots = TRUE
)## Using all 76 stations (no date filtering)
## Calculating detection probabilities...
## Using existing DE_pred column
## Calculating system probabilities...
## Creating plots...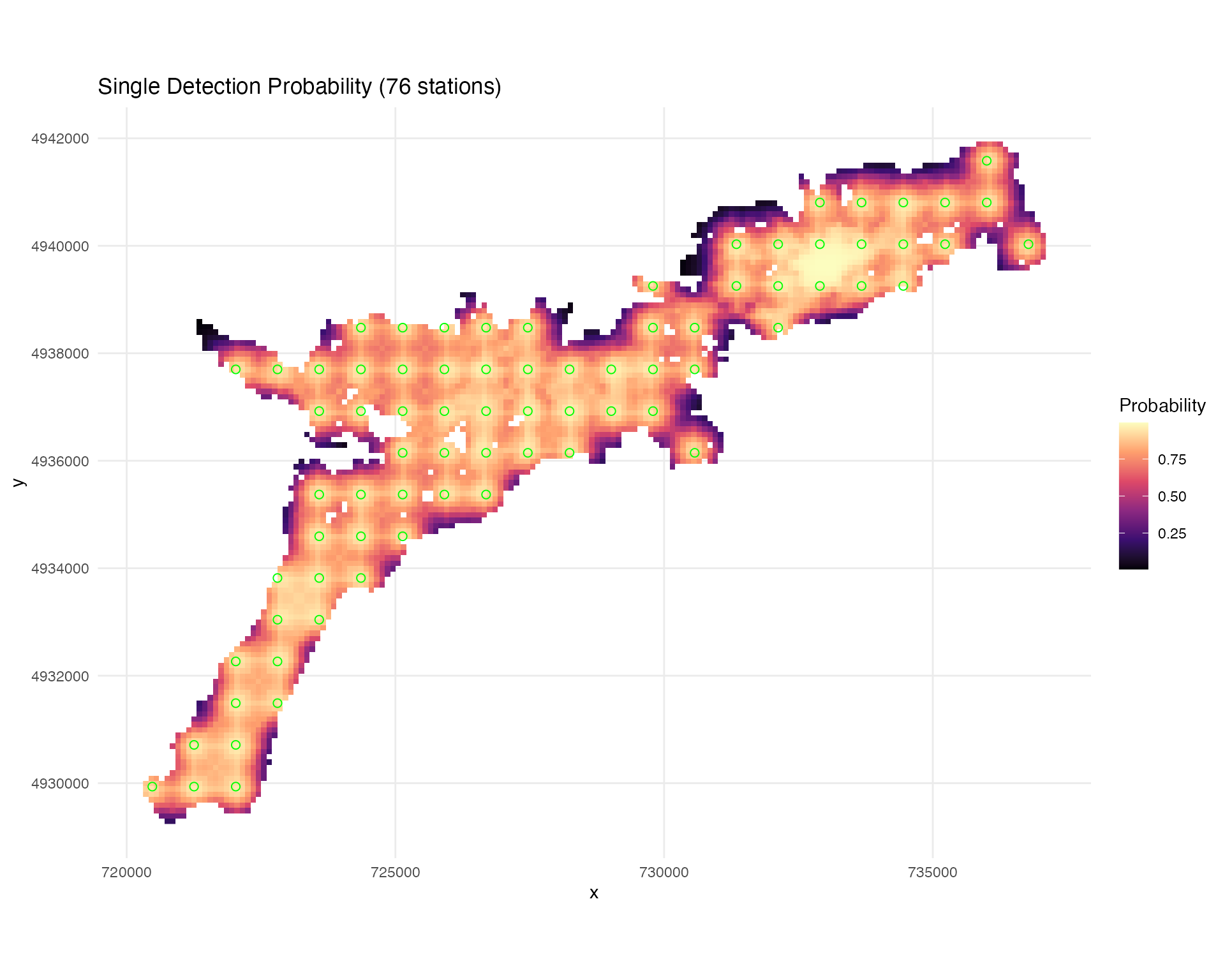
##
## === Detection System Summary ===
## Target date: All dates
## Active stations: 76
## Mean cumulative detection probability: 0.739🐟 Fish Movement Simulation
Generate Realistic Fish Tracks
# Set start time for maintaining temporal structure
start_time <- as.POSIXct("2025-07-15 12:00:00", tz = "UTC")
# Simulate correlated random walks with detection events
fish_simulation <- simulate_fish_tracks(
raster = depth_raster,
station_distances = station_distances, # Contains DE_pred values
n_paths = 5, # Number of fish to simulate
n_steps = 480, # Steps per fish (480 * 3 min = 24 hours)
step_length_mean = 50, # Mean step length (meters)
step_length_sd = 30, # Step length standard deviation
time_step = 180, # Time between steps (seconds)
seed = 123,
start_time = start_time
)## Preprocessing spatial lookup table...
## Generating path 1 of 5
## Generating path 2 of 5
## Generating path 3 of 5
## Generating path 4 of 5
## Generating path 5 of 5
## Simulation complete!
## Start time: 2025-07-15 12:00:00 UTC
## End time: 2025-07-16 12:00:00 UTCVisualize Tracks and Detections
# Plot fish tracks with detection events
plot_fish_tracks(
fish_simulation,
depth_raster,
points_regular,
show_detections = TRUE
) +
ggtitle("Simulated Fish Tracks with Detection Events",
subtitle = "Lines = movement paths, Points = detections, Triangles = receivers") +
theme_minimal()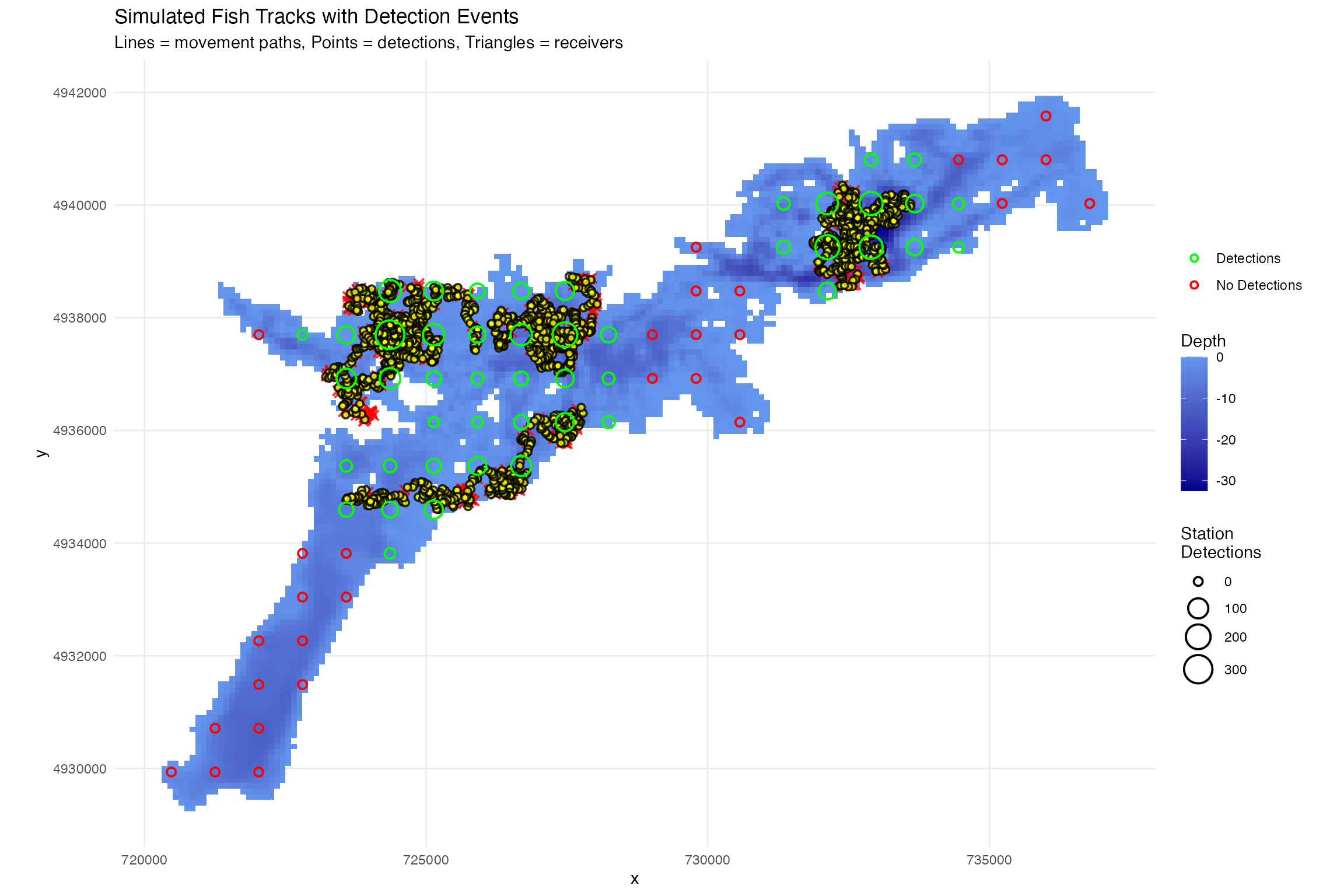
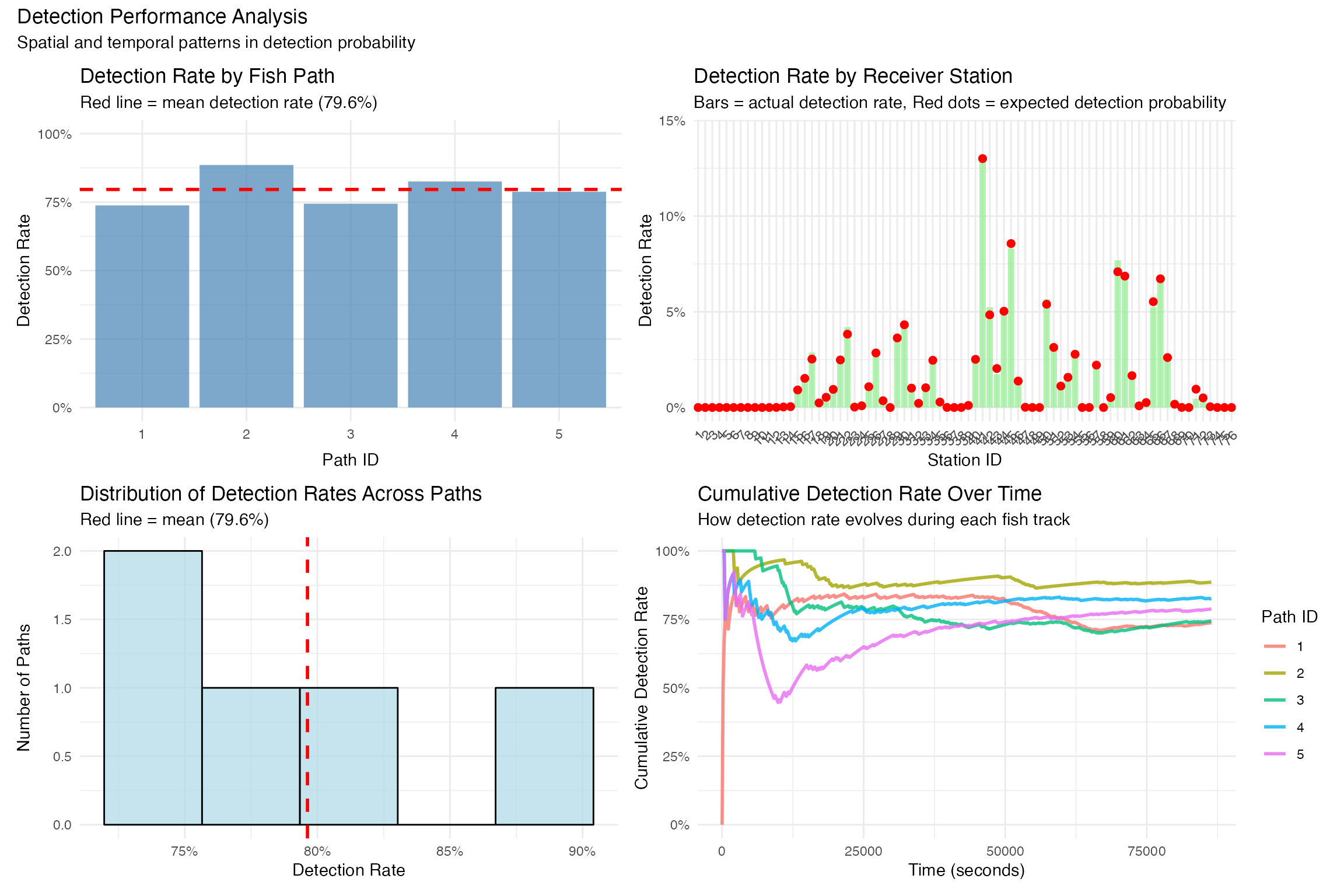
Analyze Detection Performance
# Comprehensive detection performance analysis
detection_performance <- analyze_detection_performance(fish_simulation,
create_plots = TRUE,
display_plots = FALSE)## Analyzing detection performance...
##
## === DETECTION SUMMARY REPORT ===
##
## OVERALL DETECTION PERFORMANCE:
## Total steps simulated: 2405
## Steps with detections: 1915
## Overall detection rate: 79.6%
##
## DETECTION RATE STATISTICS ACROSS PATHS:
## Mean detection rate: 79.6%
## Median detection rate: 78.8%
## Range: 73.8% - 88.6%
## Standard deviation: 6.1%
##
## DETECTION RATE BY INDIVIDUAL PATH:
## Path 1: 355/481 steps detected (73.8%)
## Path 2: 426/481 steps detected (88.6%)
## Path 3: 358/481 steps detected (74.4%)
## Path 4: 397/481 steps detected (82.5%)
## Path 5: 379/481 steps detected (78.8%)
##
## DETECTION RATE BY RECEIVER STATION:
## Station 1: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 2: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 3: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 4: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 5: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 6: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 7: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 8: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 9: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 10: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 11: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 12: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 13: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 14: 2/2405 opportunities detected (0.1%, expected 0.0%)
## Station 15: 24/2405 opportunities detected (1.0%, expected 0.9%)
## Station 16: 39/2405 opportunities detected (1.6%, expected 1.5%)
## Station 17: 69/2405 opportunities detected (2.9%, expected 2.5%)
## Station 18: 6/2405 opportunities detected (0.2%, expected 0.2%)
## Station 19: 12/2405 opportunities detected (0.5%, expected 0.5%)
## Station 20: 23/2405 opportunities detected (1.0%, expected 0.9%)
## Station 21: 61/2405 opportunities detected (2.5%, expected 2.5%)
## Station 22: 101/2405 opportunities detected (4.2%, expected 3.8%)
## Station 23: 1/2405 opportunities detected (0.0%, expected 0.0%)
## Station 24: 2/2405 opportunities detected (0.1%, expected 0.1%)
## Station 25: 25/2405 opportunities detected (1.0%, expected 1.1%)
## Station 26: 66/2405 opportunities detected (2.7%, expected 2.8%)
## Station 27: 8/2405 opportunities detected (0.3%, expected 0.4%)
## Station 28: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 29: 90/2405 opportunities detected (3.7%, expected 3.6%)
## Station 30: 108/2405 opportunities detected (4.5%, expected 4.3%)
## Station 31: 27/2405 opportunities detected (1.1%, expected 1.0%)
## Station 32: 7/2405 opportunities detected (0.3%, expected 0.2%)
## Station 33: 21/2405 opportunities detected (0.9%, expected 1.0%)
## Station 34: 60/2405 opportunities detected (2.5%, expected 2.5%)
## Station 35: 7/2405 opportunities detected (0.3%, expected 0.3%)
## Station 36: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 37: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 38: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 39: 1/2405 opportunities detected (0.0%, expected 0.1%)
## Station 40: 58/2405 opportunities detected (2.4%, expected 2.5%)
## Station 41: 311/2405 opportunities detected (12.9%, expected 13.0%)
## Station 42: 126/2405 opportunities detected (5.2%, expected 4.8%)
## Station 43: 42/2405 opportunities detected (1.7%, expected 2.0%)
## Station 44: 122/2405 opportunities detected (5.1%, expected 5.0%)
## Station 45: 200/2405 opportunities detected (8.3%, expected 8.6%)
## Station 46: 35/2405 opportunities detected (1.5%, expected 1.4%)
## Station 47: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 48: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 49: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 50: 136/2405 opportunities detected (5.7%, expected 5.4%)
## Station 51: 77/2405 opportunities detected (3.2%, expected 3.1%)
## Station 52: 28/2405 opportunities detected (1.2%, expected 1.1%)
## Station 53: 43/2405 opportunities detected (1.8%, expected 1.6%)
## Station 54: 69/2405 opportunities detected (2.9%, expected 2.8%)
## Station 55: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 56: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 57: 49/2405 opportunities detected (2.0%, expected 2.2%)
## Station 58: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 59: 13/2405 opportunities detected (0.5%, expected 0.5%)
## Station 60: 185/2405 opportunities detected (7.7%, expected 7.1%)
## Station 61: 165/2405 opportunities detected (6.9%, expected 6.9%)
## Station 62: 43/2405 opportunities detected (1.8%, expected 1.7%)
## Station 63: 2/2405 opportunities detected (0.1%, expected 0.1%)
## Station 64: 10/2405 opportunities detected (0.4%, expected 0.3%)
## Station 65: 126/2405 opportunities detected (5.2%, expected 5.5%)
## Station 66: 160/2405 opportunities detected (6.7%, expected 6.7%)
## Station 67: 58/2405 opportunities detected (2.4%, expected 2.6%)
## Station 68: 4/2405 opportunities detected (0.2%, expected 0.2%)
## Station 69: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 70: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 71: 11/2405 opportunities detected (0.5%, expected 1.0%)
## Station 72: 12/2405 opportunities detected (0.5%, expected 0.5%)
## Station 73: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 74: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 75: 0/2405 opportunities detected (0.0%, expected 0.0%)
## Station 76: 0/2405 opportunities detected (0.0%, expected 0.0%)
##
## Creating visualization plots...
# Display performance plots in grid
performance_plots <- (detection_performance$plots$by_path +
detection_performance$plots$by_station) /
(detection_performance$plots$distribution +
detection_performance$plots$time_series)
performance_plots +
plot_annotation(
title = "Detection Performance Analysis",
subtitle = "Spatial and temporal patterns in detection probability"
)
🎯 WADE Position Estimation
Calculate Position Probabilities
# Apply WADE algorithm to estimate fish positions
positioning_results <- calculate_fish_positions(
station_detections = fish_simulation$station_detections,
station_distances_df = station_distances,
station_info = points_regular,
de_model = logistic_DE$log_model,
detection_weight = 0.5, # Weight for detection evidence
non_detection_weight = 0.5, # Weight for non-detection evidence
max_non_detection_distance = 2000, # Maximum range for non-detection inference
weighting_method = "normalize_stations", # Normalize across stations
# weighting_method = "information_theoretic", # Alternative option
normalization_method = "min_max", # Min-max normalization
fish_id_col = "path_id",
time_col = "datetime",
time_aggregation = "day", # Daily position estimates
station_col = "station_id",
verbose = TRUE
)## Converting sf object to station_info data frame...
## Using station_info with static station locations and DE predictions...
## Timezone info: Input timezone = UTC , processing in UTC
## Creating receiver_stations from station_info coordinates...
## Auto-detected CRS: EPSG:32617 (UTM Zone 17N) based on coordinate ranges
## Created receiver_stations with 76 stations from station_info
## === CALCULATING FISH POSITIONS ===
## Step 1: Standardizing columns and creating time periods...
## Step 2: Aggregating detection data...
## Step 3: Creating non-detection data...
## Step 4: Aggregating non-detection data...
## Step 5: Applying normalize_stations weighting...
## Using station normalization for detections, raw DE for non-detections
## Step 6: Combining detection and non-detection data...
## Step 7: Calculating position probabilities...
## Step 8: Adding time metadata and station coordinates...
## === POSITIONING COMPLETE ===
## Fish tracked: 5
## Time periods: 2
## Time aggregation: day
## Spatial cells: 4818
## Position estimates generated: 48180Visualize Position Estimates
# Select example fish and time for visualization
fish_select <- 1
time_select <- "2025-07-15"
# Create three-panel plot showing different probability surfaces
p1 <- plot_fish_positions(
positioning_results = positioning_results,
depth_raster_df = depth_raster,
fish_select = fish_select,
time_select = time_select,
plot_type = "detection",
prob_threshold = 0.01, # Show cells above 1% probability
track_data = fish_simulation$tracks
) + ggtitle("Detection-Based Positioning",
subtitle = "Probability surface from detected stations")
p2 <- plot_fish_positions(
positioning_results = positioning_results,
depth_raster_df = depth_raster,
fish_select = fish_select,
time_select = time_select,
plot_type = "non_detection",
track_data = fish_simulation$tracks
) + ggtitle("Non-Detection-Based Positioning",
subtitle = "Inference from silent stations")
p3 <- plot_fish_positions(
positioning_results = positioning_results,
depth_raster_df = depth_raster,
fish_select = fish_select,
time_select = time_select,
plot_type = "integrated",
prob_threshold = 0.4,
detection_threshold = 0.01, # Filter to detection area
track_data = fish_simulation$tracks
) + ggtitle("Integrated Position Estimate",
subtitle = "Combined detection and non-detection evidence")
# Display all three plots
p1 + p2 + p3 + plot_spacer() + plot_layout(ncol=2)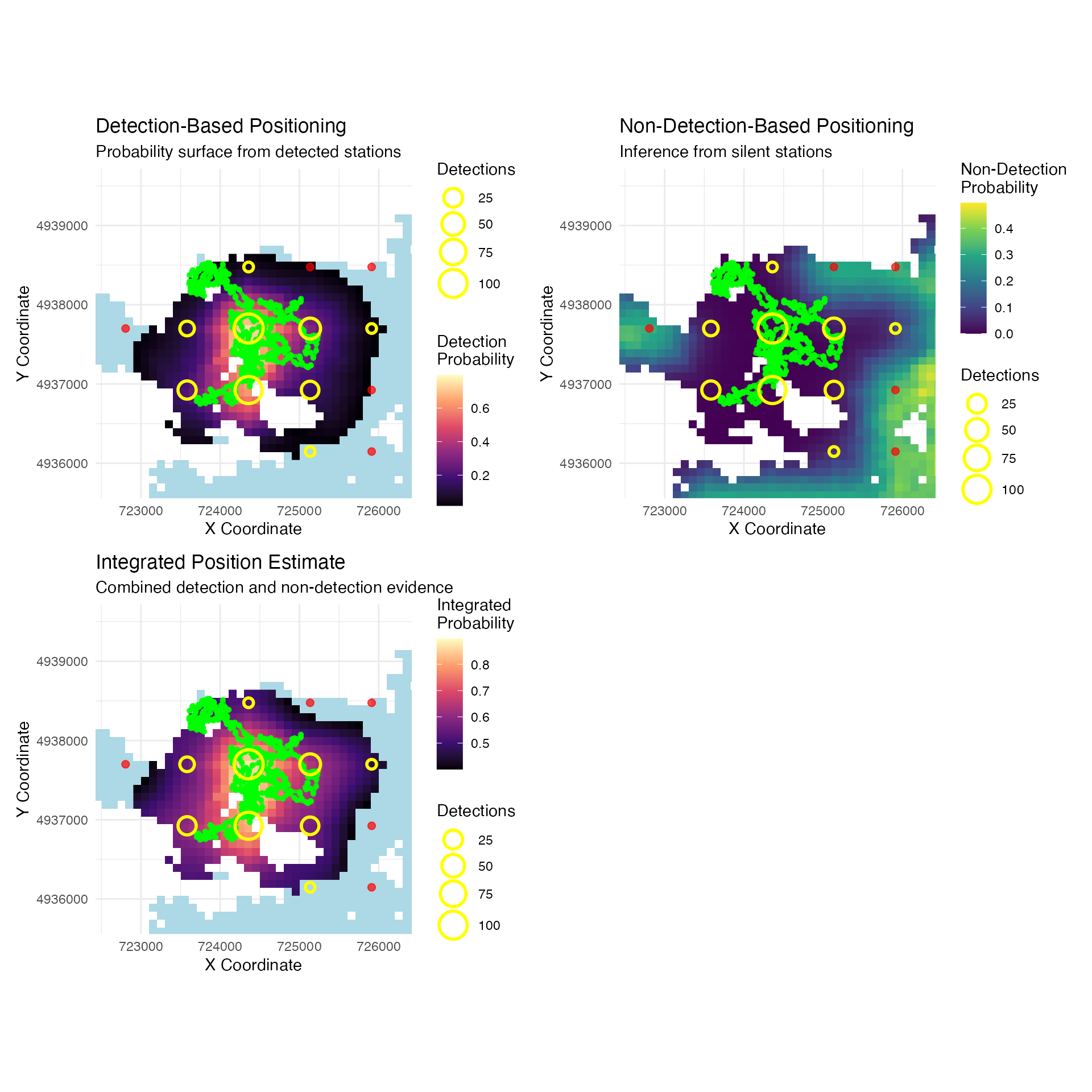
🏡 Space Use Analysis
Generate Position Points for Analysis
# Sample points from position probability surfaces
# Initially without threshold for optimization analysis
position_points <- sample_points_from_probabilities(
positioning_results,
prob_column = "weighted_mean_DE_normalized_scaled",
n_points = 1000,
min_prob_threshold = 0.0, # No threshold initially
crs = 32617,
by_group = TRUE, # Sample per fish-time combination
seed = 456
)## Using probability column: weighted_mean_DE_normalized_scaled
## Original data: 48180 cells
## After probability thresholds ( 0 < prob < 1 ): 29538 cells
##
## Processing 10 fish-time combinations
## Fish 1 - Time 2025-07-15 : sampled 1000 points from 2909 cells
## Fish 1 - Time 2025-07-16 : sampled 1000 points from 2793 cells
## Fish 2 - Time 2025-07-15 : sampled 1000 points from 2417 cells
## Fish 2 - Time 2025-07-16 : sampled 1000 points from 2418 cells
## Fish 3 - Time 2025-07-15 : sampled 1000 points from 3469 cells
## Fish 3 - Time 2025-07-16 : sampled 1000 points from 3249 cells
## Fish 4 - Time 2025-07-15 : sampled 1000 points from 3350 cells
## Fish 4 - Time 2025-07-16 : sampled 1000 points from 3408 cells
## Fish 5 - Time 2025-07-15 : sampled 1000 points from 2554 cells
## Fish 5 - Time 2025-07-16 : sampled 1000 points from 2971 cells
##
## === SAMPLING SUMMARY ===
## Total points sampled: 10000
## Probability column: weighted_mean_DE_normalized_scaled
## Fish ID(s): 1, 2, 3, 4, 5
## Time periods: 2
## Fish-time combinations: 10
## Sampled probability range: 0 to 0.9898
## Mean probability: 0.1308
# Extract coordinates from sf object
position_points_coords <- position_points %>%
dplyr::mutate(
coords = sf::st_coordinates(geometry),
x = coords[,1],
y = coords[,2]
) %>%
sf::st_drop_geometry() %>%
dplyr::select(-coords)Initial Space Use Estimates
options(positionR.verbose = FALSE)
# Calculate positioning-based space use with initial threshold
position_space_use_results <- calculate_space_use(
track_data = position_points_coords,
prob_column = "probability",
prob_thresholds = 0.2, # Initial test threshold
by_fish = TRUE,
by_time_period = TRUE,
time_aggregation = "day",
methods = c("constrained_convex_hull"),
grid_resolution = 100,
reference_raster = depth_raster
)
# Calculate actual track-based space use (ground truth)
track_space_use_results <- calculate_space_use(
track_data = fish_simulation$tracks,
by_fish = TRUE,
by_time_period = TRUE,
time_aggregation = "day",
methods = c("grid_cell_count", "constrained_convex_hull"),
grid_resolution = 100,
reference_raster = depth_raster
)Visual Comparison of Space Use
# Compare positioning vs track space use
fish_select <- 1
time_select <- "2025-07-15"
p1 <- plot_space_use(
position_space_use_results,
plot_type = "map",
fish_select = fish_select,
time_select = time_select,
method_select = "constrained_convex_hull",
track_data = fish_simulation$tracks,
background_raster = depth_raster
) + ggtitle("Constrained Convex Hull Space Use (Positioning)")
p2 <- plot_space_use(
track_space_use_results,
plot_type = "map",
track_data = fish_simulation$tracks,
fish_select = fish_select,
time_select = time_select,
method_select = "constrained_convex_hull",
background_raster = depth_raster
) + ggtitle("Constrained Convex Hull Space Use (Track)",
subtitle = "Ground truth from actual movement")
p1 | p2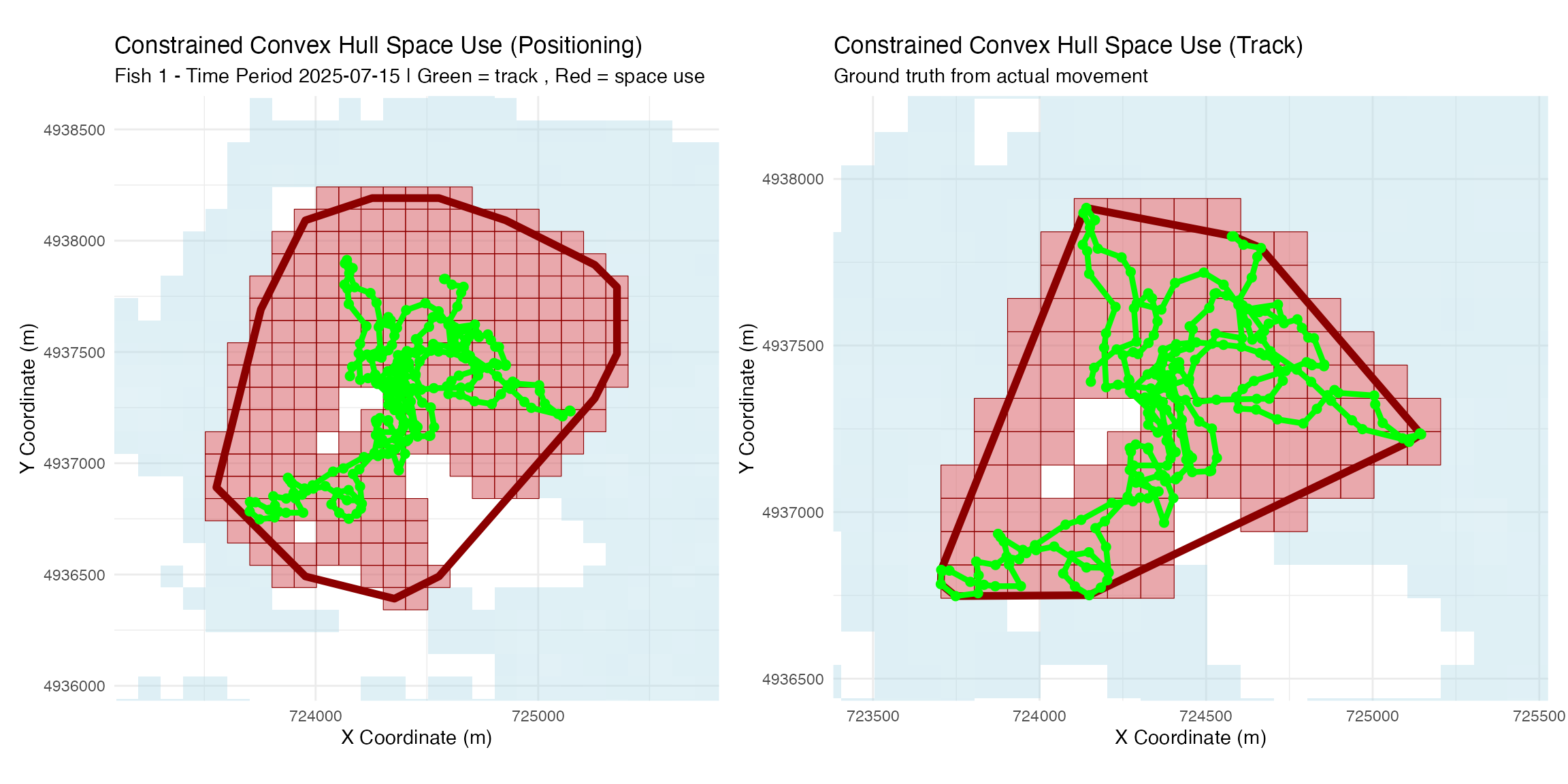
🎚️ Probability Threshold Optimization
Multi-Threshold Analysis
options(positionR.verbose = FALSE)
# Test multiple probability thresholds
multi_results <- calculate_space_use(
track_data = position_points_coords,
prob_column = "probability",
by_fish = TRUE,
by_time_period = TRUE,
prob_thresholds = c(0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6),
methods = c("constrained_convex_hull"),
reference_raster = depth_raster
)
# Compare against track-based space use
# Note: Track overlap calculation may have issues
comparison <- compare_space_use_thresholds(
multi_threshold_results = multi_results,
reference_tracks = fish_simulation$tracks,
reference_space_use = track_space_use_results
)## === SPACE USE THRESHOLD COMPARISON ===
## Analyzing 7 thresholds: 0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6
## Analyzing methods: constrained_convex_hull
##
## Processing threshold: 0
## Method: constrained_convex_hull
##
## Processing threshold: 0.1
## Method: constrained_convex_hull
##
## Processing threshold: 0.2
## Method: constrained_convex_hull
##
## Processing threshold: 0.3
## Method: constrained_convex_hull
##
## Processing threshold: 0.4
## Method: constrained_convex_hull
##
## Processing threshold: 0.5
## Method: constrained_convex_hull
##
## Processing threshold: 0.6
## Method: constrained_convex_hull
##
## Creating visualization plots...
##
## === THRESHOLD COMPARISON COMPLETE ===
## Thresholds analyzed: 7
## Methods analyzed: 1
## Fish-time combinations: 9.714286
# Display optimization results
print(comparison$plots$area_ratio_boxplot) 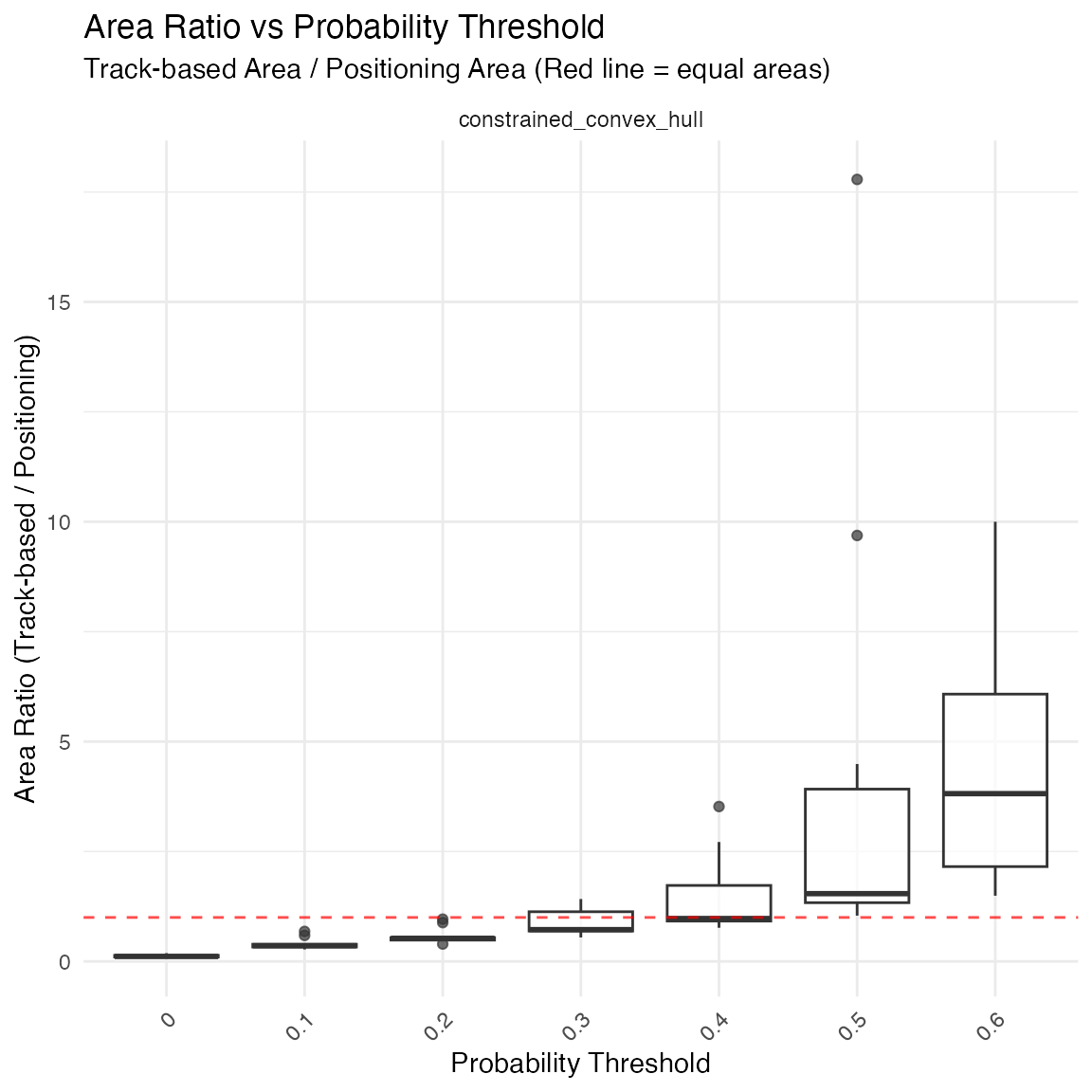
print(comparison$plots$track_overlap_boxplot)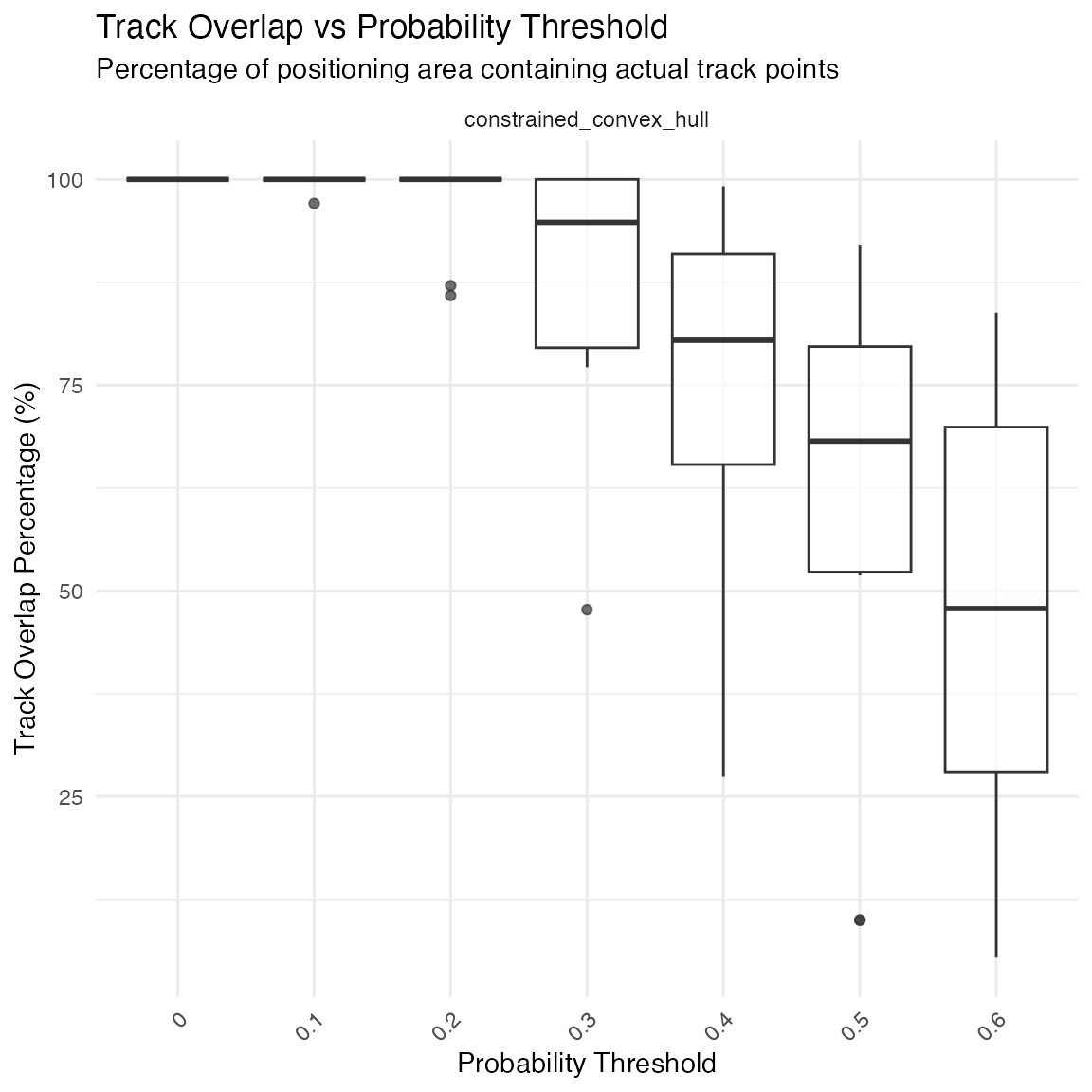
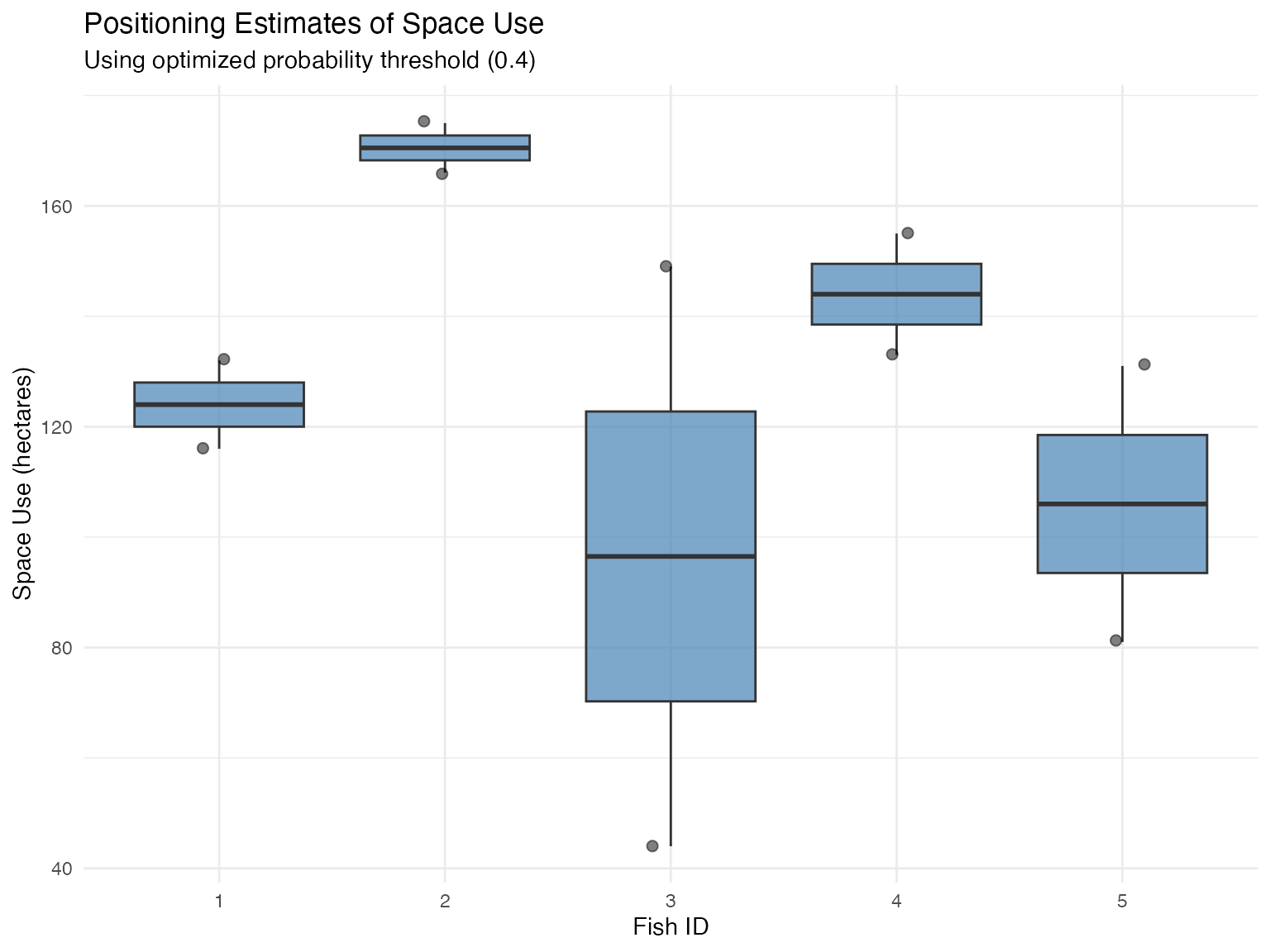
Final Space Use with Optimal Threshold
# Calculate space use with optimized threshold
space_use_0.4 <- calculate_space_use(
track_data = position_points_coords,
prob_column = "probability",
by_fish = TRUE,
by_time_period = TRUE,
prob_thresholds = 0.4, # Optimal threshold from analysis
methods = c("constrained_convex_hull"),
reference_raster = depth_raster
)
# Visualize final space use estimates
ggplot(space_use_0.4$space_use_estimates,
aes(as.factor(fish_id), constrained_convex_hull_area_hectares)) +
geom_boxplot(fill = "steelblue", alpha = 0.7) +
geom_jitter(width = 0.1, alpha = 0.5, size = 2) +
theme_minimal() +
labs(
title = "Positioning Estimates of Space Use",
subtitle = "Using optimized probability threshold (0.4)",
x = "Fish ID",
y = "Space Use (hectares)"
)
# Summary statistics
space_summary <- space_use_0.4$space_use_estimates %>%
group_by(fish_id) %>%
summarise(
mean_area = mean(constrained_convex_hull_area_hectares),
sd_area = sd(constrained_convex_hull_area_hectares),
.groups = 'drop'
)🌿 Habitat Selection Analysis
Generate Presence Points for Habitat Analysis
# Note: Need to integrate filtering approaches from field applications
# Generate presence points using optimal threshold
position_points <- sample_points_from_probabilities(
positioning_results,
prob_column = "weighted_mean_DE_normalized_scaled",
n_points = 1000,
min_prob_threshold = 0.4, # Based on optimization results
crs = 32617,
by_group = TRUE,
seed = 456
)## Using probability column: weighted_mean_DE_normalized_scaled
## Original data: 48180 cells
## After probability thresholds ( 0.4 < prob < 1 ): 1038 cells
##
## Processing 10 fish-time combinations
## Fish 1 - Time 2025-07-15 : sampled 1000 points from 101 cells
## Fish 1 - Time 2025-07-16 : sampled 1000 points from 111 cells
## Fish 2 - Time 2025-07-15 : sampled 1000 points from 150 cells
## Fish 2 - Time 2025-07-16 : sampled 1000 points from 153 cells
## Fish 3 - Time 2025-07-15 : sampled 1000 points from 120 cells
## Fish 3 - Time 2025-07-16 : sampled 1000 points from 37 cells
## Fish 4 - Time 2025-07-15 : sampled 1000 points from 129 cells
## Fish 4 - Time 2025-07-16 : sampled 1000 points from 113 cells
## Fish 5 - Time 2025-07-15 : sampled 1000 points from 55 cells
## Fish 5 - Time 2025-07-16 : sampled 1000 points from 69 cells
##
## === SAMPLING SUMMARY ===
## Total points sampled: 10000
## Probability column: weighted_mean_DE_normalized_scaled
## Fish ID(s): 1, 2, 3, 4, 5
## Time periods: 2
## Fish-time combinations: 10
## Sampled probability range: 0.4006 to 0.9898
## Mean probability: 0.5565
# Visualize presence points
raster_df <- as.data.frame(depth_raster, xy = TRUE)
ggplot() +
geom_raster(data = raster_df, aes(x = x, y = y, fill = layer)) +
scale_fill_gradient(low = "blue4", high = "cornflowerblue",
na.value = "transparent", name = "Depth (m)") +
geom_sf(data = position_points %>%
filter(fish_id == 1 & time_period_label == "2025-07-15"),
aes(color = probability),
alpha = 0.5, size = 1) +
scale_color_viridis_c(name = "Probability", option = "magma") +
theme_minimal() +
labs(title = "Presence Points",
subtitle = "Fish 1, July 15 - Points weighted by position probability")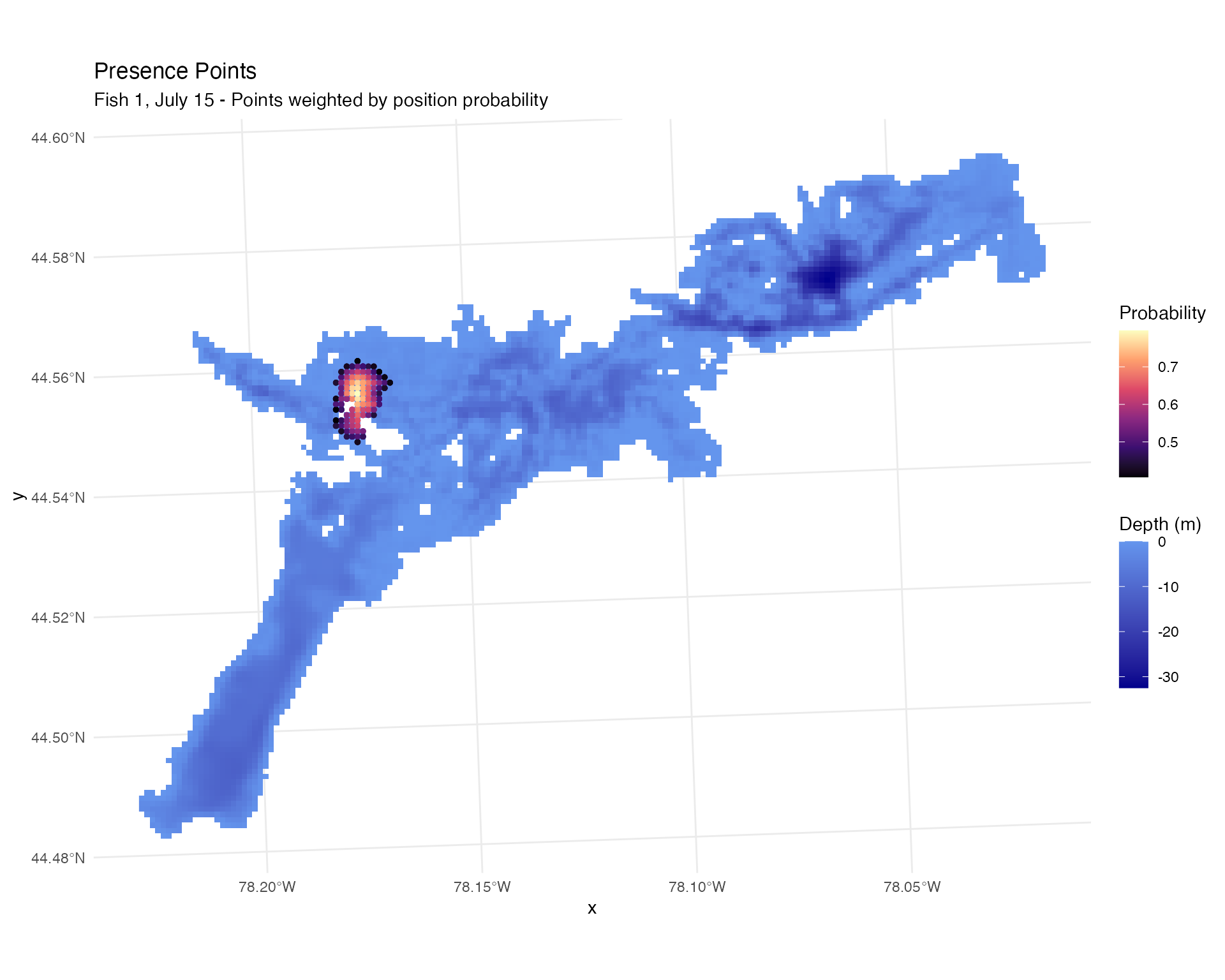
Generate Absence Points
System DE-Weighted Absences
# Generate absences weighted by system detection efficiency
absence_points <- sample_points_from_system_de(
system_DE,
position_points = position_points,
n_points = 1000,
min_prob_threshold = 0.05,
crs = 32617,
seed = 123
)## Using probability column: cumulative_prob
## Original data: 4818 cells
## Found 5 fish ID(s) in position_points template
## Found 2 time period(s) in position_points template
## Note: Template contains fish/time information, but system_de data lacks these columns.
## Will replicate sampling across template groups.## After probability thresholds ( 0.05 < prob < 1 ): 4798 cells
##
## Replicating sampling across template fish-time combinations
## Found 10 unique fish-time combinations in template
## fish_id = 1, time_period = 1752537600, time_period_posix = 1752537600, time_period_label = 2025-07-15 : replicated 1000 points
## fish_id = 1, time_period = 1752624000, time_period_posix = 1752624000, time_period_label = 2025-07-16 : replicated 1000 points
## fish_id = 2, time_period = 1752537600, time_period_posix = 1752537600, time_period_label = 2025-07-15 : replicated 1000 points
## fish_id = 2, time_period = 1752624000, time_period_posix = 1752624000, time_period_label = 2025-07-16 : replicated 1000 points
## fish_id = 3, time_period = 1752537600, time_period_posix = 1752537600, time_period_label = 2025-07-15 : replicated 1000 points
## fish_id = 3, time_period = 1752624000, time_period_posix = 1752624000, time_period_label = 2025-07-16 : replicated 1000 points
## fish_id = 4, time_period = 1752537600, time_period_posix = 1752537600, time_period_label = 2025-07-15 : replicated 1000 points
## fish_id = 4, time_period = 1752624000, time_period_posix = 1752624000, time_period_label = 2025-07-16 : replicated 1000 points
## fish_id = 5, time_period = 1752537600, time_period_posix = 1752537600, time_period_label = 2025-07-15 : replicated 1000 points
## fish_id = 5, time_period = 1752624000, time_period_posix = 1752624000, time_period_label = 2025-07-16 : replicated 1000 points
##
## === SAMPLING SUMMARY ===
## Total points sampled: 10000
## Probability column: cumulative_prob
## Groups: 10
## Sampled probability range: 0.0978 to 0.9948
## Mean probability: 0.7923
ggplot() +
geom_raster(data = raster_df, aes(x = x, y = y, fill = layer)) +
scale_fill_gradient(low = "blue4", high = "cornflowerblue",
na.value = "transparent", name = "Depth (m)") +
geom_sf(data = absence_points %>%
filter(fish_id == 1 & time_period_label == "2025-07-15"),
aes(color = probability),
alpha = 1, size = 0.8) +
scale_color_viridis_c(name = "Cumulative\nProbability") +
theme_minimal() +
labs(title = "Absence Points (System DE Weighted)",
subtitle = "Sampling probability proportional to detection coverage")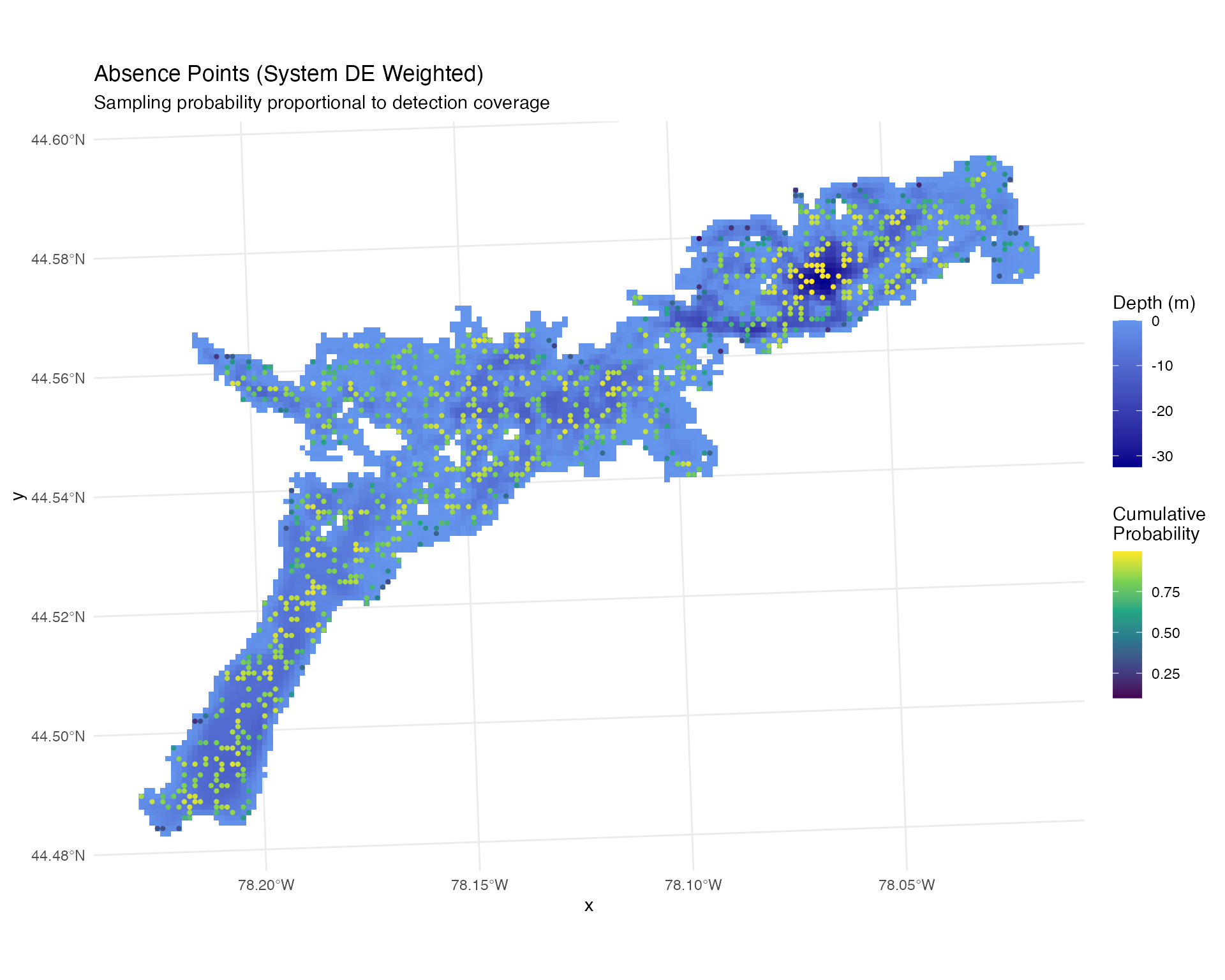
Uniform Random Absences
# Generate uniform absences across entire system
absence_points_uniform <- sample_points_from_system_de(
system_DE,
position_points = position_points,
n_points = 1000,
uniform = TRUE, # Uniform sampling
min_prob_threshold = 0.0, # No threshold - whole system
crs = 32617,
seed = 123
)## Using probability column: cumulative_prob
## Original data: 4818 cells
## Found 5 fish ID(s) in position_points template
## Found 2 time period(s) in position_points template
## Note: Template contains fish/time information, but system_de data lacks these columns.
## Will replicate sampling across template groups.## After probability thresholds ( 0 < prob < 1 ): 4818 cells
##
## Replicating sampling across template fish-time combinations
## Found 10 unique fish-time combinations in template
## fish_id = 1, time_period = 1752537600, time_period_posix = 1752537600, time_period_label = 2025-07-15 : replicated 1000 points
## fish_id = 1, time_period = 1752624000, time_period_posix = 1752624000, time_period_label = 2025-07-16 : replicated 1000 points
## fish_id = 2, time_period = 1752537600, time_period_posix = 1752537600, time_period_label = 2025-07-15 : replicated 1000 points
## fish_id = 2, time_period = 1752624000, time_period_posix = 1752624000, time_period_label = 2025-07-16 : replicated 1000 points
## fish_id = 3, time_period = 1752537600, time_period_posix = 1752537600, time_period_label = 2025-07-15 : replicated 1000 points
## fish_id = 3, time_period = 1752624000, time_period_posix = 1752624000, time_period_label = 2025-07-16 : replicated 1000 points
## fish_id = 4, time_period = 1752537600, time_period_posix = 1752537600, time_period_label = 2025-07-15 : replicated 1000 points
## fish_id = 4, time_period = 1752624000, time_period_posix = 1752624000, time_period_label = 2025-07-16 : replicated 1000 points
## fish_id = 5, time_period = 1752537600, time_period_posix = 1752537600, time_period_label = 2025-07-15 : replicated 1000 points
## fish_id = 5, time_period = 1752624000, time_period_posix = 1752624000, time_period_label = 2025-07-16 : replicated 1000 points
##
## === SAMPLING SUMMARY ===
## Total points sampled: 10000
## Probability column: cumulative_prob
## Groups: 10
## Sampled probability range: 0.0109 to 0.9903
## Mean probability: 0.74
ggplot() +
geom_raster(data = raster_df, aes(x = x, y = y, fill = layer)) +
scale_fill_gradient(low = "blue4", high = "cornflowerblue",
na.value = "transparent", name = "Depth (m)") +
geom_sf(data = absence_points_uniform %>%
filter(fish_id == 1 & time_period_label == "2025-07-15"),
alpha = 1, size = 0.8, col = "red") +
theme_minimal() +
labs(title = "Absence Points (Uniform Across System)",
subtitle = "Random sampling across entire study area")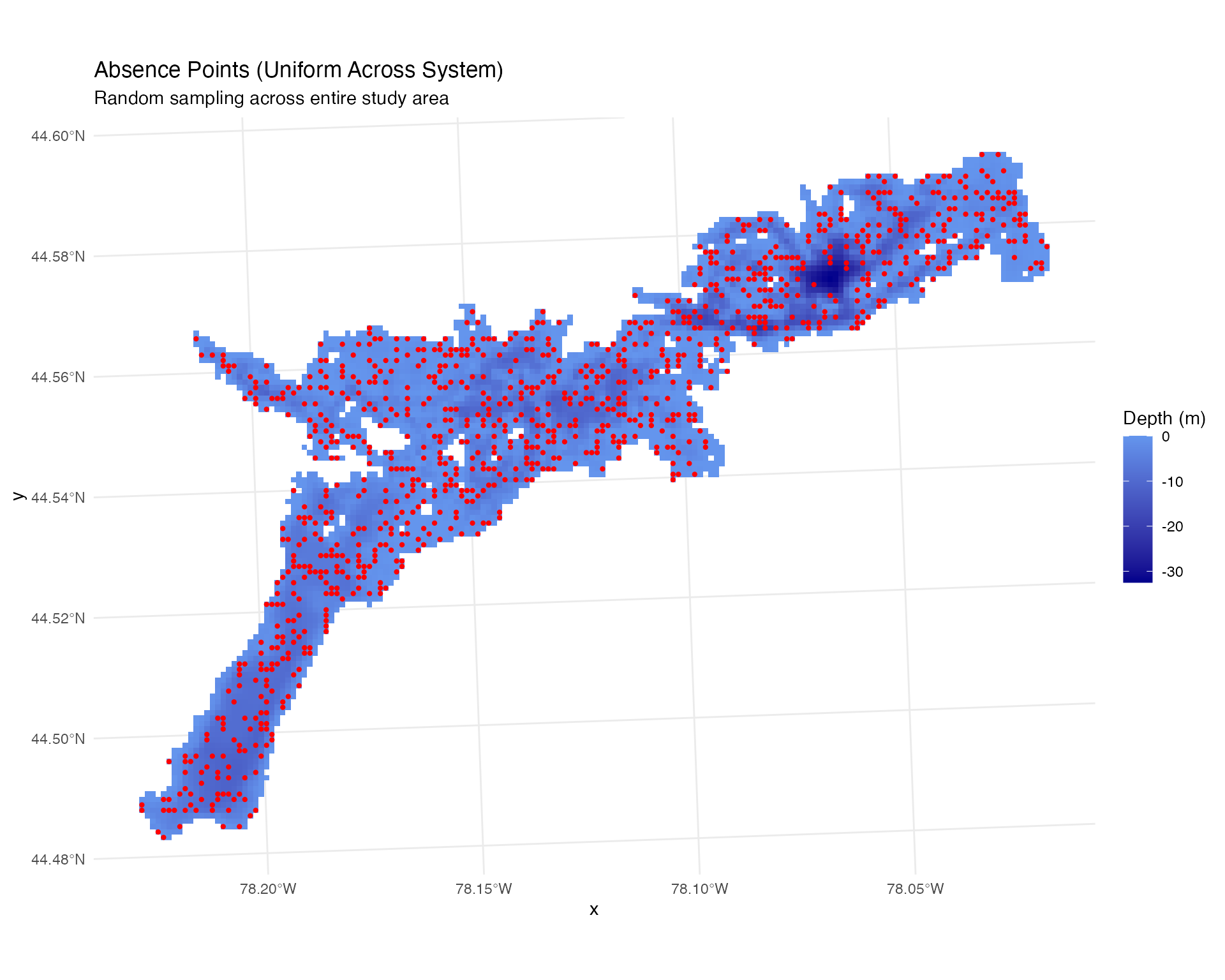
Depth Selection Analysis - Positioning
# Extract depth values
position_points$depth_m <- raster::extract(depth_raster, position_points)
absence_points_uniform$depth_m <- raster::extract(depth_raster, absence_points_uniform)
# Combine presence and absence data
# Using uniform absences for consistency with track analysis
presence_absence_points <- rbind(
position_points %>%
select(fish_id, time_period_posix, depth_m) %>%
mutate(type = "presence"),
absence_points_uniform %>%
select(fish_id, time_period_posix, depth_m) %>%
mutate(type = "absence")
)
# Create depth selection plots
p1 <- plot_depth_selection(presence_absence_points, plot_type = "density") +
ggtitle("Depth Selection",
subtitle = "Blue = presences, Red = absences")## Plotting depth selection for 20000 points
p2 <- plot_depth_comparison(
presence_absence_points,
comparison_var = "fish_id",
plot_type = "boxplot"
) +
ggtitle("Depth Selection by Individual Fish")
p1 | p2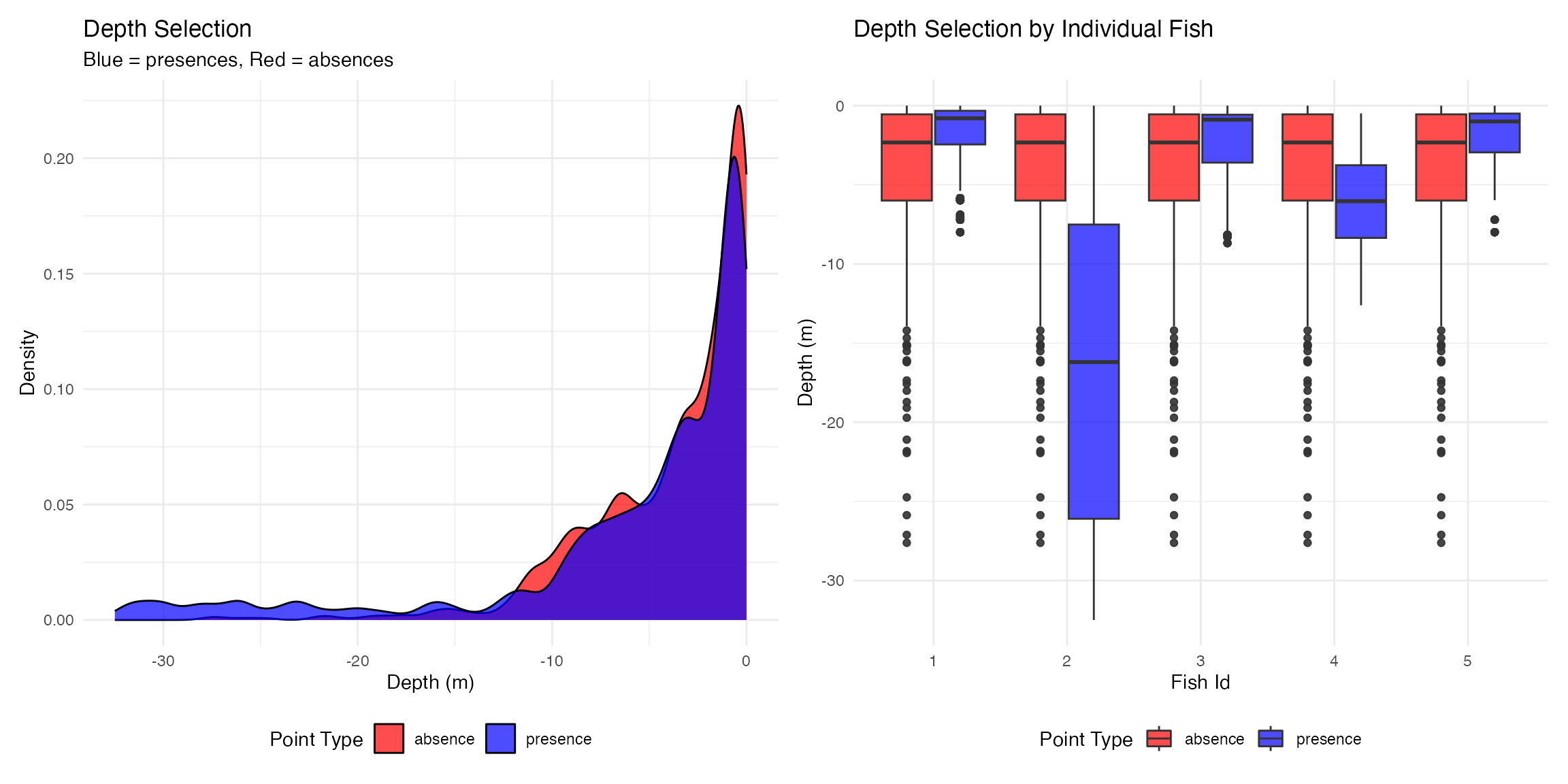
🐠 Track-Based Habitat Selection (Ground Truth)
Generate Track Presence Points
# Sample points from grid cells used by actual tracks
track_presence <- sample_points_from_track_grid(
track_data = fish_simulation$tracks,
reference_raster = depth_raster,
n_points = 1000,
crs = 32617
)## === TRACK GRID SAMPLING ===
## Grid resolution: 100 meters
## Original track data: 2405 points
## Creating grid and counting track points...
## Created 787 grid cells with track points
## After count thresholds ( 1 <=count<= Inf ): 787 cells
##
## Processing 10 fish-time combinations
## fish_id = 1, time_period_label = 2025-07-15, time_period_posix = 1752537600 : sampled 1000 points from 67 cells (total usage: 240 track points)
## fish_id = 1, time_period_label = 2025-07-16, time_period_posix = 1752624000 : sampled 1000 points from 62 cells (total usage: 241 track points)
## fish_id = 2, time_period_label = 2025-07-15, time_period_posix = 1752537600 : sampled 1000 points from 80 cells (total usage: 240 track points)
## fish_id = 2, time_period_label = 2025-07-16, time_period_posix = 1752624000 : sampled 1000 points from 86 cells (total usage: 241 track points)
## fish_id = 3, time_period_label = 2025-07-15, time_period_posix = 1752537600 : sampled 1000 points from 78 cells (total usage: 240 track points)
## fish_id = 3, time_period_label = 2025-07-16, time_period_posix = 1752624000 : sampled 1000 points from 79 cells (total usage: 241 track points)
## fish_id = 4, time_period_label = 2025-07-15, time_period_posix = 1752537600 : sampled 1000 points from 83 cells (total usage: 240 track points)
## fish_id = 4, time_period_label = 2025-07-16, time_period_posix = 1752624000 : sampled 1000 points from 83 cells (total usage: 241 track points)
## fish_id = 5, time_period_label = 2025-07-15, time_period_posix = 1752537600 : sampled 1000 points from 85 cells (total usage: 240 track points)
## fish_id = 5, time_period_label = 2025-07-16, time_period_posix = 1752624000 : sampled 1000 points from 84 cells (total usage: 241 track points)
##
## === SAMPLING SUMMARY ===
## Total points sampled: 10000
## Grid resolution: 100 meters
## Fish ID(s): 1, 2, 3, 4, 5
## Time periods: 2
## Groups: 10
## Usage range: 1 to 15 track points per cell
## Mean usage: 4.64 track points per cell
ggplot() +
geom_raster(data = raster_df, aes(x = x, y = y, fill = layer)) +
scale_fill_gradient(low = "blue4", high = "cornflowerblue",
na.value = "transparent", name = "Depth (m)") +
geom_sf(data = track_presence,
aes(color = count),
alpha = 0.5, size = 1) +
scale_color_viridis_c(name = "Count", option = "magma") +
coord_sf() +
theme_minimal() +
labs(title = "Track Presence Points",
subtitle = "Sampled from actual movement paths")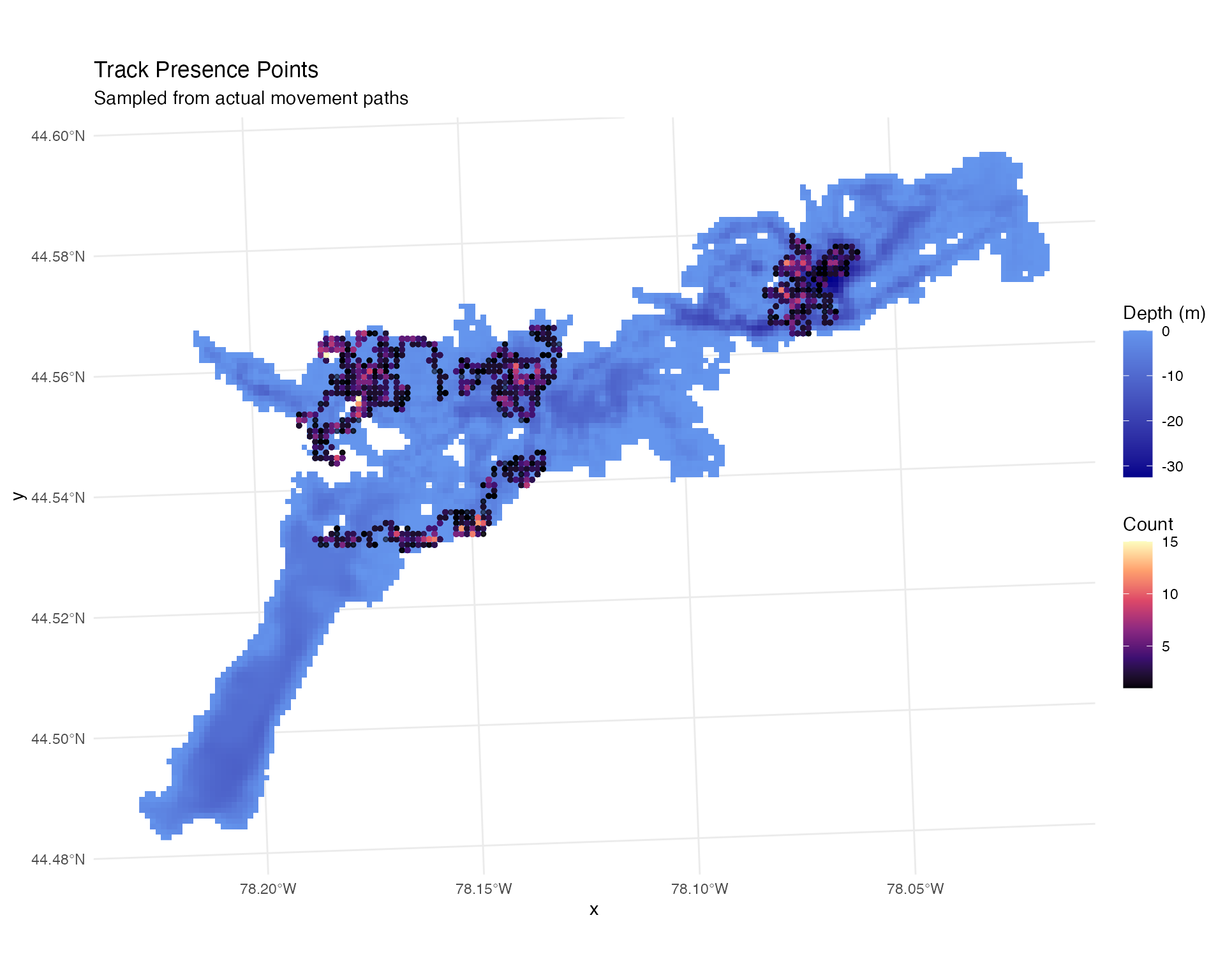
Generate Track Absence Points
# Generate absences for track-based analysis
track_absence <- sample_points_from_system_de(
system_DE,
position_points = track_presence,
n_points = 1000,
uniform = TRUE, # Uniform sampling
min_prob_threshold = 0.0, # Whole system
crs = 32617,
seed = 123
)## Using probability column: cumulative_prob
## Original data: 4818 cells
## Found 5 fish ID(s) in position_points template
## Found 2 time label(s) in position_points template
## Note: Template contains fish/time information, but system_de data lacks these columns.
## Will replicate sampling across template groups.## After probability thresholds ( 0 < prob < 1 ): 4818 cells
##
## Replicating sampling across template fish-time combinations
## Found 10 unique fish-time combinations in template
## fish_id = 1, time_period_posix = 1752537600, time_period_label = 2025-07-15 : replicated 1000 points
## fish_id = 1, time_period_posix = 1752624000, time_period_label = 2025-07-16 : replicated 1000 points
## fish_id = 2, time_period_posix = 1752537600, time_period_label = 2025-07-15 : replicated 1000 points
## fish_id = 2, time_period_posix = 1752624000, time_period_label = 2025-07-16 : replicated 1000 points
## fish_id = 3, time_period_posix = 1752537600, time_period_label = 2025-07-15 : replicated 1000 points
## fish_id = 3, time_period_posix = 1752624000, time_period_label = 2025-07-16 : replicated 1000 points
## fish_id = 4, time_period_posix = 1752537600, time_period_label = 2025-07-15 : replicated 1000 points
## fish_id = 4, time_period_posix = 1752624000, time_period_label = 2025-07-16 : replicated 1000 points
## fish_id = 5, time_period_posix = 1752537600, time_period_label = 2025-07-15 : replicated 1000 points
## fish_id = 5, time_period_posix = 1752624000, time_period_label = 2025-07-16 : replicated 1000 points
##
## === SAMPLING SUMMARY ===
## Total points sampled: 10000
## Probability column: cumulative_prob
## Groups: 10
## Sampled probability range: 0.0109 to 0.9903
## Mean probability: 0.74
ggplot() +
geom_raster(data = raster_df, aes(x = x, y = y, fill = layer)) +
scale_fill_gradient(low = "blue4", high = "cornflowerblue",
na.value = "transparent", name = "Depth (m)") +
geom_sf(data = track_absence %>%
filter(fish_id == 1 & time_period_label == "2025-07-15"),
alpha = 1, size = 0.8, col = "red") +
theme_minimal() +
labs(title = "Track Absence Points (Uniform Across System)",
subtitle = "Random background points for comparison")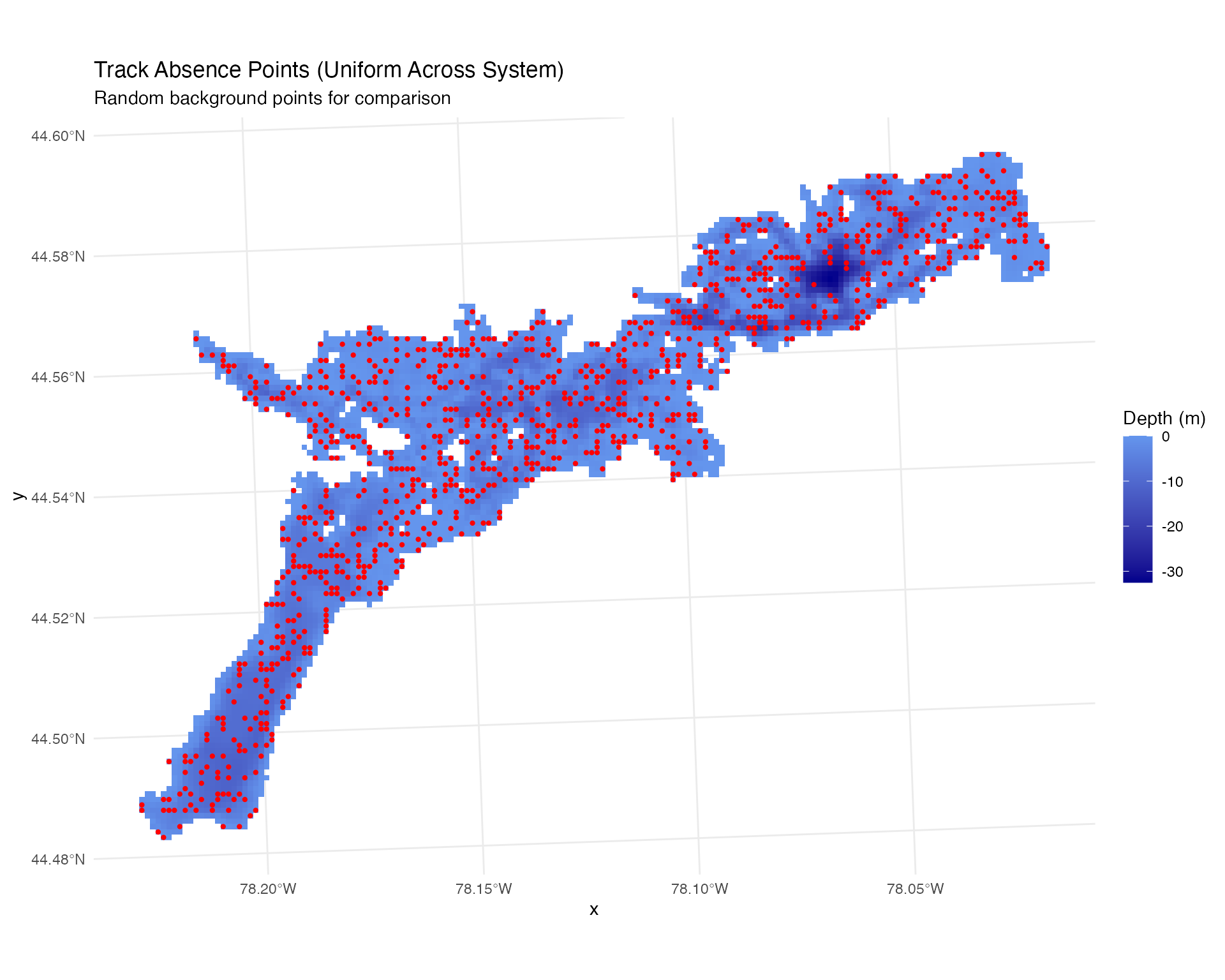
Depth Selection Analysis - Tracks
# Extract depth values for track data
track_presence$depth_m <- raster::extract(depth_raster, track_presence)
track_absence$depth_m <- raster::extract(depth_raster, track_absence)
# Combine track presence and absence data
track_presence_absence_points <- rbind(
track_presence %>%
select(fish_id, time_period_posix, depth_m) %>%
mutate(type = "presence"),
track_absence %>%
select(fish_id, time_period_posix, depth_m) %>%
mutate(type = "absence")
)
# Create track depth selection plots
p1 <- plot_depth_selection(track_presence_absence_points, plot_type = "density") +
ggtitle("Depth Selection (Track-Based Ground Truth)",
subtitle = "Blue = presences, Red = absences")## Plotting depth selection for 20000 points
p2 <- plot_depth_comparison(
track_presence_absence_points,
comparison_var = "fish_id",
plot_type = "boxplot"
) +
ggtitle("Depth Selection by Individual Fish (Tracks)")
p1 | p2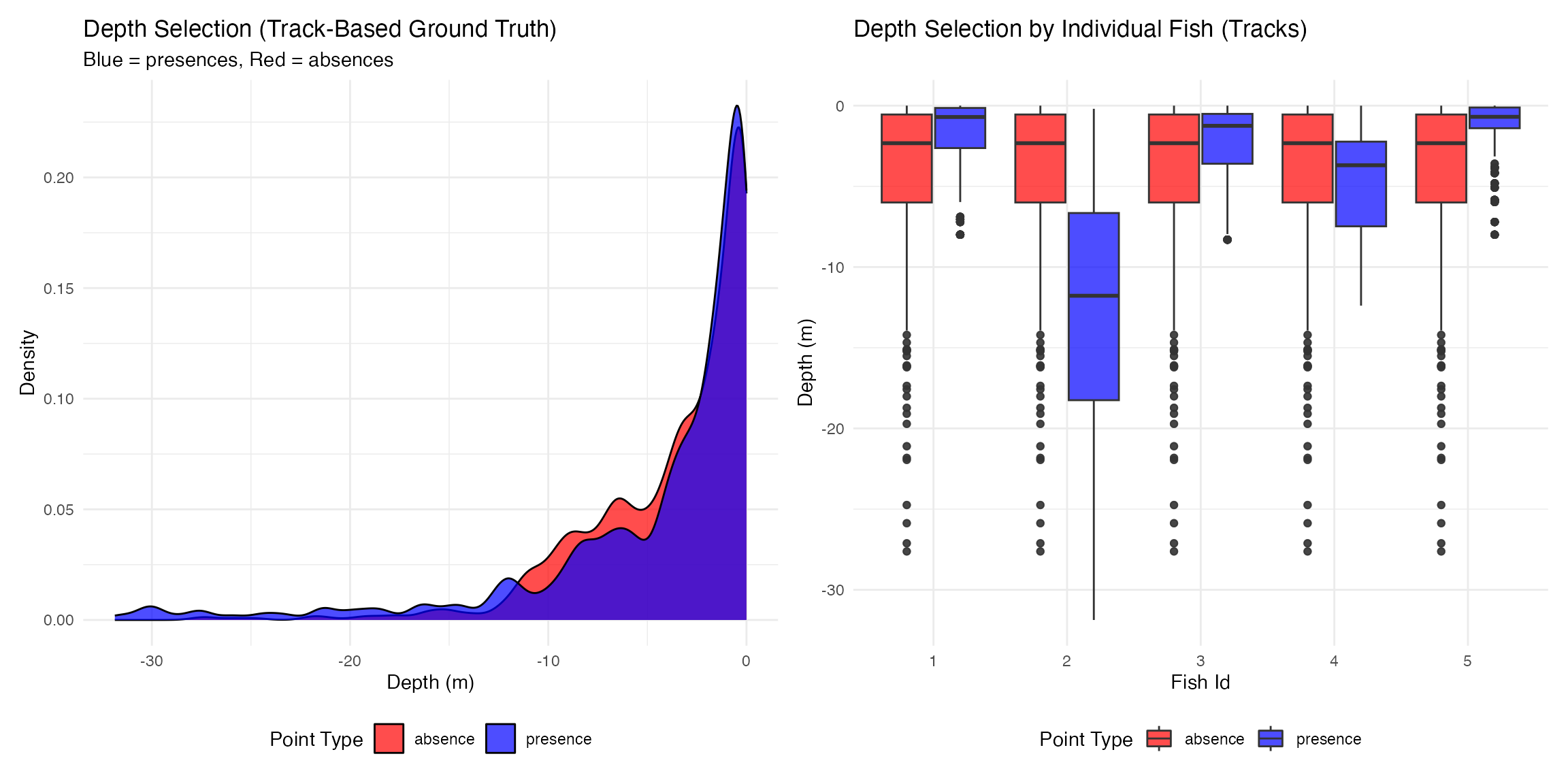
🔬 Comparative Habitat Selection Analysis
Random Forest Model Comparison
# Comprehensive comparison using Random Forests
comparative_results <- analyze_comparative_habitat_selection(
positioning_data = presence_absence_points,
space_use_data = track_presence_absence_points,
analysis_type = "both",
formula = presence ~ depth_m + fish_id + time_period_posix,
time_variable = "time_period_posix",
sample_size = 10000,
ntree = 500,
create_plots = TRUE,
create_comparison = TRUE
)## === COMPARATIVE HABITAT SELECTION ANALYSIS ===
##
## --- ANALYZING POSITIONING-BASED DATA ---
## Positioning data - Original: 20000 Complete: 20000
## Sampled 10000 points for Positioning model
## Fitting Positioning random forest model...
## Positioning Model OOB Error: 32.97 %
##
## --- ANALYZING SPACE USE-BASED DATA ---
## Space Use data - Original: 20000 Complete: 19954
## Sampled 10000 points for Space Use model
## Fitting Space Use random forest model...
## Space Use Model OOB Error: 32.83 %
##
## --- CREATING COMPARISON PLOTS ---
##
## === COMPARATIVE ANALYSIS COMPLETE ===
# Display comprehensive summary
cat("=== COMPARATIVE HABITAT SELECTION ANALYSIS ===\n\n")## === COMPARATIVE HABITAT SELECTION ANALYSIS ===
print_comparative_summary(comparative_results)## === COMPARATIVE HABITAT SELECTION SUMMARY ===
## Analysis Type: both
## Datasets Analyzed: positioning, space_use, comparison
##
## --- POSITIONING RESULTS ---
## OOB Error Rate: 32.97 %
## Sample Size Used: 10000
## Depth Range: -32.5 0
## Top Variable (MeanDecreaseGini): depth_m ( 386.055 )
## Probability Range: 0.357 0.566
## Mean Probability: 0.479
##
## --- SPACE_USE RESULTS ---
## OOB Error Rate: 32.83 %
## Sample Size Used: 10000
## Depth Range: -31.87 0
## Top Variable (MeanDecreaseGini): depth_m ( 302.935 )
## Probability Range: 0.271 0.563
## Mean Probability: 0.445
##
## --- COMPARISON STATISTICS ---
## Mean Difference (Pos - Space): 0.0321
## Mean Absolute Difference: 0.0379
## Maximum Difference: 0.1139
## Correlation: 0.8841
##
## === SUMMARY COMPLETE ===Visualize Comparative Results
# Plot all comparative results
plot_comparative_results(comparative_results, plot_type = "comparison")##
## Comparison plots: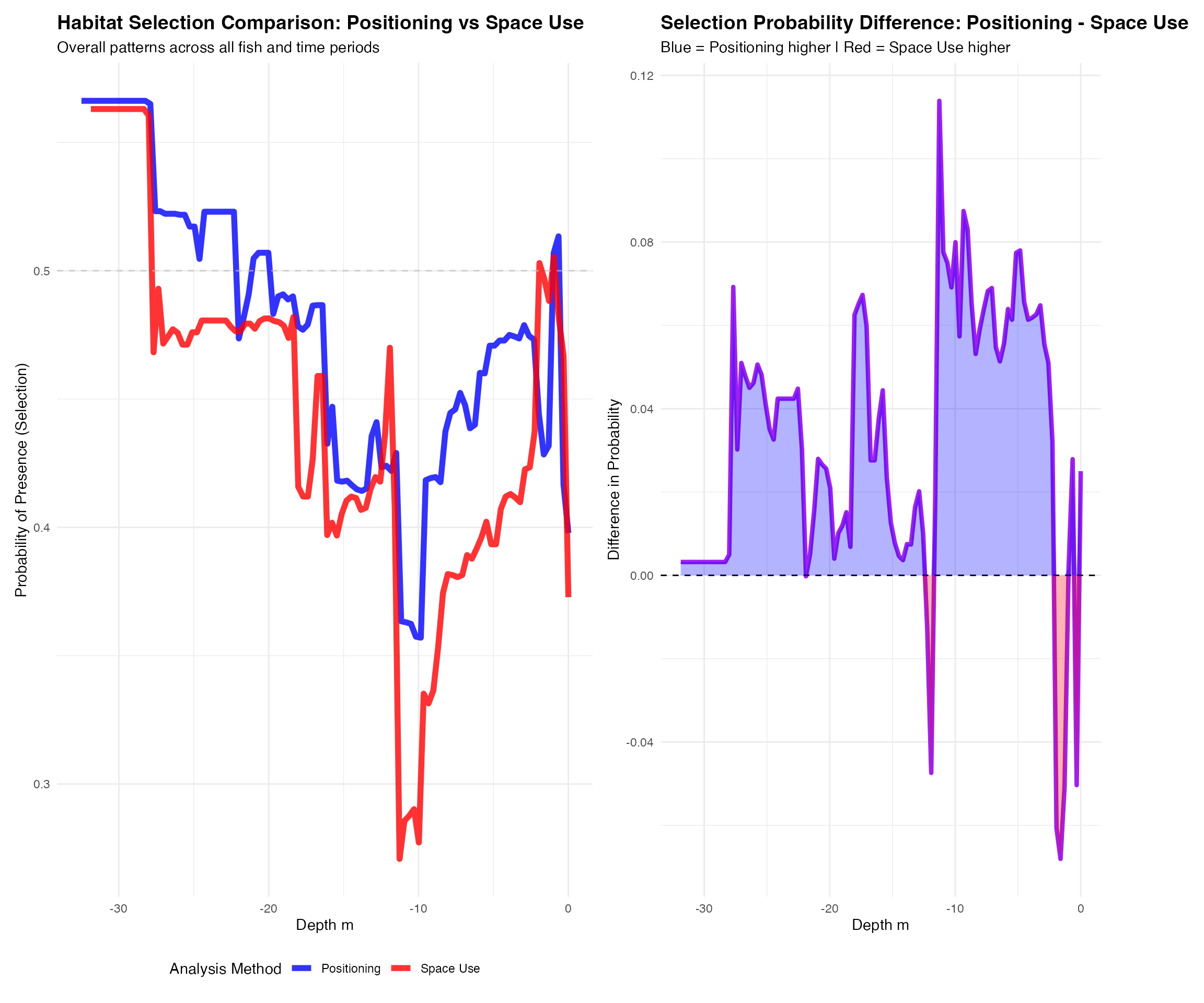
📋 Summary & Conclusions
Key Takeaways
Here we’ve used a simulation based dataset to conduct WADE positioning, which produces useful outputs for assessing space use (home range) and habitat selection from acoustic telemetry data.
There are a variety of options for the WADE algorithm in how the data are processed (e.g., normalizing station detection ranges, weighting of detections vs non-detecting receivers, threshold cut offs for positioning). Conducting simulation-based analysis within your system can aid in determining the optimal approaches relative to system design, detection efficiency, and animal behaviour by comparing the accuracy of positioning outputs with the known track data.
Tools for generating reasonable detection range models that integrate depth are provided, although it is much more optimal to use real detection range data and models to inform this. Detection range varies considerably over space and time due to variation in environmental conditions.